Abstract
To promote the clinical and epidemiological studies that improve our understanding of human genetic susceptibility to environmental exposure, the Environmental Genome Project (EGP) has scanned 213 environmental response genes involved in DNA repair, cell cycle regulation, apoptosis, and metabolism for single nucleotide polymorphisms (SNPs). Many of these genes have been implicated by loss-of-function mutations associated with severe diseases attributable to decreased protection of genomic integrity. Therefore, the hypothesis for these studies is that individuals with functionally significant polymorphisms within these genes may be particularly susceptible to genotoxic environmental agents. On average, 20.4 kb of baseline genomic sequence or 86% of each gene, including a substantial amount of introns, all exons, and 1.3 kb upstream and downstream, were scanned for variations in the 90 samples of the Polymorphism Discovery Resource panel. The average nucleotide diversity across the 4.2 MB of these 213 genes is 6.7 × 10-4, or one SNP every 1500 bp, when two random chromosomes are compared. The average candidate environmental response gene contains 26 PHASE inferred haplotypes, 34 common SNPs, 6.2 coding SNPs (cSNPs), and 2.5 nonsynonymous cSNPs. SIFT and Polyphen analysis of 541 nonsynonymous cSNPs identified 57 potentially deleterious SNPs. An additional eight polymorphisms predict altered protein translation. Because these genes represent 1% of all known human genes, extrapolation from these data predicts the total genomic set of cSNPs, nonsynonymous cSNPs, and potentially deleterious nonsynonymous cSNPs. The implications for the use of these data in direct and indirect association studies of environmentally induced diseases are discussed.
The link between environmental agents and disease risk has been recognized for more than a century with the discovery of the link between coal soot and scrotal cancer in young chimney sweeps (Doll 1975). Increased disease risk due to a combination of a specific genetic background and exposure to environmental agents has been known since protein polymorphisms could be associated with disease phenotypes. Early examples are hemolysis from antimalarial drugs and other oxidants in individuals with glucose-6-phosphate dehydrogenase deficiency (Motulsky 1972), increased risk of emphysema from cigarette smoking in individuals with alpha1-antitrypsin deficiency (Lieberman et al. 1969; Hutchison et al. 1970), and lactose intolerance in individuals with lactase deficiency (Dahlqvist et al. 1963; Klotz 1964; Haemmerli et al. 1965). To explore the role of common genetic polymorphisms in environmentally induced disease in United States populations, the National Institute of Environmental Health Sciences (NIEHS) initiated the Environmental Genome Project (EGP) in 1997 (Olden and Wilson 2000).
The initial efforts of the EGP have focused on the discovery and annotation of single nucleotide polymorphisms (SNPs), the most common form of human genetic variation, in candidate environmental disease genes and the development of databases integrating sequence polymorphism data into individually annotated human and mouse gene models (GeneSNPs, http://www.genome.utah.edu/genesnps; PolyDom, http://polydoms.cchmc.org; Trafac, http://genometrafac.cchmc.org). These efforts began with 213 candidate environmental susceptibility genes from a list of 550 candidates submitted based on their involvement in processes influenced by environmental exposure. Broadly grouped into pathways related to cell cycle, cell signaling, cell structure, DNA repair, gene expression, and metabolism, many of these genes have been implicated by loss-of-function mutations associated with severe diseases attributable to decreased protection of genomic integrity. Thus, the hypothesis for these studies is that individuals with functionally significant polymorphisms within these genes may be particularly susceptible to genotoxic environmental agents. For example, the tumor suppressor genes RB1, ATM, and MLH1 are associated with familial cancers and may also harbor minor risk alleles that may conspire with exposure to environmental agents to diminish the fidelity of DNA repair and increase the lifetime risk of developing cancer (Alonso et al. 2001; Mathonnet et al. 2003; Buchholz et al. 2004).
The first phase of the EGP is now complete, and herein we report the discovery of 23,443 SNPs in 213 candidate environmental response genes by systematically resequencing DNA samples from 90 individuals of the polymorphism discovery resource (PDR) panel. The PDR panel is a representative panel of individuals drawn from the United States population, including Americans of European, African, Mexican, and Asian descent and Native Americans. The explicit objective of the panel is to facilitate detection of polymorphic sites that occur in any one of the represented populations (Collins et al. 1998). The PDR panel of 90 samples was used because this sample size has excellent chances for detecting polymorphic sites occurring at >5% minor allele frequency (MAF) in any one of the ethnic subpopulations (Kruglyak and Nickerson 2001). Because this panel was designed to discover human genetic variation while being sensitive to the ethical, legal, and social issues of population definition, and not to assess the frequency of variations in ethnic subpopulations, all identifying demographic information was removed from the individual samples.
The results of this analysis, cataloged in the GeneSNPs and dbSNP databases, suggest the presence of a significant number of polymorphisms that may confer sensitivity to environmental agents, and are stimulating ongoing efforts to (1) develop mouse models of potentially functional polymorphisms (Comparative Mouse Genomics Centers Consortium); (2) explore the common variant-common disease hypothesis via molecular epidemiology studies of environmentally induced diseases; (3) address the ethical, legal, and social implications (ELSIs) of the genetics of environmental disease susceptibility; and (4) improve strategies for the discovery of genetic variations responsible for the elevated sensitivity to environmental agents.
RESULTS
Candidate Genes
Comprehensive polymorphism discovery was performed by resequencing 213 candidate environmental response genes involved in DNA repair (70), apoptosis (41), cell cycle control (62), and drug metabolism (40). These genes are distributed across all the human chromosomes except for the Y chromosome, and altogether represent slightly <1% of all known human genes (Ewing and Green 2000; Lander et al. 2001). On average, ∼86% of the genomic sequence for each gene was scanned for variation across 90 DNA samples from the PDR (Collins et al. 1998). Genomic sequencing coverage ranged from 100% coverage for 38 genes to 14% for E2F3. Notably, 32 of the 213 candidate genes (15%) have already been implicated in Mendelian diseases, including 21 disease genes for rare forms of cancer susceptibility, such as the breast cancer susceptibility locus BRCA1, the retinoblastoma locus RB1, and the Wilms tumor locus WT1 (see Supplemental Table 1). Of the 213 genes scanned, 132 (62%) were nearly completely scanned for polymorphism by resequencing >75% of the entire reference gene sequence (5′, coding, noncoding [intronic], and 3′ regions). More than 50% of the genomic sequence was resequenced in 179 of the candidate genes. The candidates ranged in size from the 0.6 kb SPHAR (S-phase responsive) to the 106 kb PMS1 (postmeiotic segregation increased 1) gene. For each candidate, all exons (316 kb total), an average of 1.3 kb upstream of the cDNA sequence (282 kb total), an average of 1.3 kb downstream of the last exon (282 kb total), and a significant amount of intronic sequence (3.26 Mb) was scanned for polymorphism discovery. In total, >4.2 Mb of baseline human reference sequence was scanned across the 90 individuals of the PDR, generating 378 Mb of sequence (the equivalent of resequencing human chromosome 13 three times). Sequence scanning led to the discovery of 23,443 SNPs and generated 2.1 million genotypes, which are cataloged at the GeneSNPs (http://genome.utah.edu/genesnps) and dbSNPs databases. Of the 23,443 SNPs, 17,538 (75%) were unique reports. Of the 17,538 new dbSNP submissions, 2928 (17%) were common, with a MAF >5%, and 14,610 (83%) had a MAF ≤5%. Of the 25% of our SNPs previously reported, only 1763 variations (29%) included allele frequency estimates.
Sequence Diversity in Candidate Genes
The overall nucleotide diversity (π) for the 213 candidate genes was 6.7 × 10-4 (equivalent to 1 SNP every 1500 bp between any two chromosomes), and the SNP frequency across the 180 chromosomes averaged one SNP every 179 bp. These estimates are consistent with previous genome-wide estimates of nucleotide diversity and SNP frequency (Li and Sadler 1991; Nickerson et al. 1998; Halushka et al. 1999; Sachidanandam et al. 2001; Stephens et al. 2001; Carlson et al. 2004). However, significant variance around this mean was observed, and nucleotide diversity varied 27-fold from 0.72 × 10-4 (equivalent to one SNP every 13,800 bp between any two chromosomes) for the MARCKS like protein (MLP) to 19.3 × 10-4 (one SNP every 500 bp between any two chromosomes) for BCL2/adenovirus E1B interacting protein 2 (BNIP2). To contrast these genes, none of the 16 variable sites in MLP had MAF >5% in the sequenced population (n = 90 samples), and therefore, none can be considered common in the population. In comparison, 198 polymorphisms in the 28-kb BNIP2 were identified, and 69 of these variable sites (34%) were common in the population, having a MAF >5%.
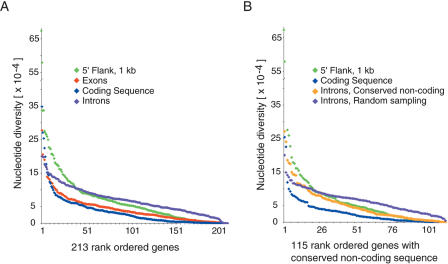
The average nucleotide diversity was classified by gene structure in the 3′ flanking region (7.0 × 10-4, ±11.7 × 10-4), intronic sequence (6.9 × 10-4, ±4.2 × 10-4), and 5′ UTR (6.9 × 10-4, ±18.0 × 10-4), and was similar to the 213-gene-wide average of 6.7 × 10-4. However, nucleotide diversity in coding regions was half that of noncoding regions, or 3.5 × 10-4 (Fig. 1A; Supplemental Table 2). Interestingly, in noncoding regions conserved in the mouse, rat, and dog, the average nucleotide diversity was intermediate (5.2 × 10-4, or one SNP every 1928 bp between any two chromosomes) between that of the coding and 5′ flanking sequence and that of the intronic sequence (Fig. 1B; Supplemental Table 3). To show that this result was not due to the difference in target size for which nucleotide diversity was being determined, we also performed a random sampling of the introns by sampling the same amount of nucleotides identified as conserved noncoding sequence (Fig. 1B).
Figure 1.
Rank order of nucleotide diversity (π) in conserved noncoding, exon, intron, CDS, and 5′ flanking regions of the EGP genes. (A) Values are calculated for all 213 EGP genes for regions containing 1 kb 5′ of the transcription initiation site (green, 174 kb), exons (red, 483 kb), coding sequence (blue, 312 kb), and introns (purple, 3.26 Mb). (B) π values are calculated for 115 EGP genes that had significant amounts of intronic conserved noncoding sequence (as described in Methods) for regions containing 1 kb 5′ of the transcription initiation site (green, 91 kb), coding sequence (blue, 189 kb) and intronic conserved noncoding (orange, 170 kb), and randomly sampled intron (purple, 2.39 Mb). All categories were sorted independently before plotting. The randomly sampled intron category corresponds to the mean π value per gene obtained by 10,000 independent samplings of the same size range as the intronic conserved noncoding region for that gene. The average SEM for this sampling was 9.3E-06 with a minimum and maximum SEM of 3.12E-07 and 3.07E-05, respectively.
A View of Sequence Diversity in the Average Candidate Gene
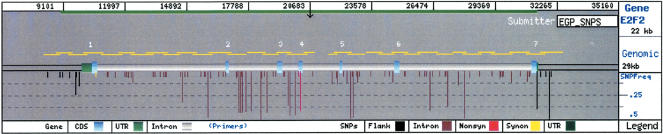
On average, 20.4 kb of reference baseline sequence was scanned for each candidate gene. The cell cycle gene E2F transcription factor 2 (E2F2) is representative of the average gene structure, sequence diversity, and size. A representation of the polymorphism distribution and gene structure of E2F2 is shown in Figure 2, and each of the candidate genes examined by the EGP is available in a similar format via the GeneSNPs database (http://genome.utah.edu/genesnps). E2F2 is coded by seven exons distributed across 21.3 kb. One hundred twelve single nucleotide substitutions and six small insertion/deletion polymorphisms were identified in this gene and are depicted by position in the gene by vertical descending bars with length that is proportional to the MAF in the study population. However, because of the masked population stratification of the PDR panel, allele frequencies within the constituent ethnic subpopulations may vary from the frequencies of the whole panel. The nucleotide diversity across E2F2 is 6.9 × 10-4, or one SNP every 1.4 kb between two random chromosomes. The number of common polymorphisms is similar to the 213-gene average, with 41% of the total (46 of 112 SNPs) having a MAF >5% in the PDR 90 panel. Also typical of the average gene, E2F2 has four cSNPs, with two nonsynonymous cSNPs indicated by the red vertical bars in Figure 2.
Figure 2.
A GeneSNPs view of E2F2 (E2F Elongation Factor 2). E2F2 represents an average of the genes scanned for polymorphism discovery based on its size and nucleotide diversity. E2F2 has seven exons (depicted by light blue rectangles for coding and green for untranslated [UTR] sequences in the mRNA). For this gene, 24 kb was scanned for polymorphisms, which includes sequences 5′ to the first exon (∼1.7 kb) and 3′ of the last exon (∼1 kb) by amplifying 90 DNA samples using 34 overlapping amplicons (horizontal yellow bars above the gene structure). Vertical descending lines indicate the position of the SNPs identified in this sequence. The length of the vertical lines represents the frequency of the minor allele, and the color indicates whether the SNP location is in flanking (black), intronic (brown), synonymous (yellow), nonsynonymous (red), or UTR (green) sequences.
Site Selection for Functional and Association Studies
In this study, the average candidate gene contains ∼34 common SNPs. Because functional analysis via animal models or genotype-phenotype studies is costly, reducing the number of sites for further analysis (from an average of 34) is a major consideration in designing effective association studies. Two complementary approaches have been proposed to identify phenotypically important SNPs. Using the direct interrogation approach focuses on testing the nonsynonymous (potentially functional) variations in coding sequence for specific phenotypes (Collins et al. 1997; Kruglyak and Nickerson 2001; Botstein and Risch 2003). Of the 23,443 SNPs found in the 213 candidate genes, 541 nonsynonymous cSNPs were identified. We evaluated these cSNPs to identify variants with potential functional consequence by using two homology-based tools: SIFT (Ng and Henikoff 2003) and Polyphen (Sunyaev et al. 2001; Ramensky et al. 2002). Fifty-seven SNPs (∼10% of total nonsynonymous SNPs) were identified as potentially deleterious by both of these approaches (Table 1). For a subset of these variants (n = 36), we were able to identify a functional domain associated with the polymorphism by using annotation from the Human Gene Mutation Database. Notably, we predict intolerant cSNPs in 31 genes with no entry in the Human Gene Mutation Database (Table 1). Seven of these predicted intolerant cSNPS have allele frequencies >5% in the PDR, four of which are not listed in the Human Gene Mutation Database and are discussed below.
Table 1.
Potentially Deleterious Nonsynonymous cSNPs Predicted by SIFT and Polyphen
| Symbola | LocusLinkb | rs (dbSNP)c | AA Pos.d | Major allelee | Minor allelef | Freqg | Pfam domainsh |
|---|---|---|---|---|---|---|---|
| ARSA | 410 | 6151411 | 82 | P | L | 0.01 | PF00884 Sulfatase |
| BRCA1 | 672 | 356 | Q | R | 0.01 | ||
| CASP2 | 835 | 4647338 | 424 | R | G | 0.02 | PF00655 ICE-like protease caspase p10 domain |
| CCNG2 | 901 | 28 | E | G | 0.01 | PF00134 cyclin | |
| CDC6 | 990 | 4135016 | 378 | R | H | 0.01 | PF01078 Mg_chelatase PF00004 AAA |
| CDC6 | 990 | 4135013 | 299 | T | M | 0.01 | PF01078 Mg_chelatasePF00004 AAA |
| CDK7 | 1022 | 285 | T | M | 0.02 | PF00069 pkinase | |
| CDKN1B | 1027 | 2066828 | 15 | R | W | 0.01 | |
| CYP2C9 | 1559 | 144 | R | C | 0.06 | PF00067 p450 | |
| CYP2C9 | 1559 | 251 | H | R | 0.01 | PF00067 p450PF03011 PFEMP | |
| CYP2C9 | 1559 | 489 | P | S | 0.01 | ||
| E2F3 | 1871 | 4134973 | 344 | G | R | 0.01 | PF02319 E2F_TDP |
| ERBB2 | 2064 | 4252633 | 452 | W | C | 0.01 | |
| ERCC1 | 2067 | 3212977 | 266 | A | T | 0.01 | PF00633 HHH |
| ERCC4 | 2072 | 1799802 | 379 | P | S | 0.01 | |
| ERCC4 | 2072 | 1800124 | 875 | E | G | 0.01 | |
| ERCC5 | 2073 | 1104 | D | H | 0.38 | ||
| ERCC5 | 2073 | 311 | S | C | 0.01 | ||
| EXO1 | 9156 | 93 | R | G | 0.01 | ||
| EXO1 | 9156 | 456 | S | Y | 0.01 | ||
| FGF11 | 2256 | 163 | R | C | 0.01 | PF00167 FGF | |
| GCKR | 2646 | 446 | P | L | 0.33 | ||
| GCKR | 2646 | 256 | G | S | 0.01 | PF01380 SIS | |
| GPI | 2821 | 208 | I | T | 0.02 | PF00342 PGI | |
| GSR | 2936 | 110 | R | C | 0.03 | PF00070 pyr_redox | |
| GSTZ1 | 2954 | 32 | E | K | 0.3 | PF02798 GST_N | |
| GTF2H1 | 2965 | 234 | R | W | 0.01 | ||
| HGF | 3082 | 5745688 | 330 | D | Y | 0.01 | PF00051 Kringle domain |
| HSPB2 | 3316 | 4252589 | 111 | G | S | 0.01 | PF00011 HSP20 |
| IGF1R | 3480 | 605 | R | H | 0.01 | ||
| IGF2R | 3482 | 203 | P | L | 0.02 | PF00878 CIMR_repeat | |
| IGF2R | 3482 | 231 | G | D | 0.01 | PF00878 CIMR_repeat | |
| LPO | 4025 | 414 | R | Q | 0.01 | PF03098 Animal haem peroxidase | |
| LPO | 4025 | 514 | R | Q | 0.01 | PF03098 Animal haem peroxidase | |
| LPO | 4025 | 614 | I | T | 0.01 | PF03098 Animal haem peroxidase | |
| MTHFR | 4524 | 429 | E | A | 0.27 | ||
| MTHFR | 4524 | 222 | A | V | 0.22 | PF02219 MTHFR | |
| MTHFR | 4524 | 422 | G | R | 0.01 | ||
| MUTYH | 4595 | 3219494 | 500 | G | E | 0.01 | |
| MYC | 4609 | 4645959 | 11 | N | S | 0.02 | PF01056 Myc_N_term |
| NAT2 | 10 | 64 | R | Q | 0.02 | PF00797 Acetyltransf2 | |
| NEIL1 | 79661 | 5745906 | 83 | G | D | 0.01 | PF01149 Fapy_DNA_glyco |
| NFKB1 | 4790 | 4648099 | 711 | H | Q | 0.02 | |
| PKMYT1 | 9088 | 140 | R | C | 0.01 | PF00069 pkinase | |
| PMS1 | 5378 | 1145232 | 501 | G | R | 0.01 | PF02465 HATPase_c |
| PMS1 | 5378 | 1145234 | 793 | Y | H | 0.01 | |
| PNKP | 11284 | 3739206 | 478 | V | G | 0.01 | |
| POLB | 5423 | 3136797 | 242 | P | R | 0.01 | PF00966 DNA_polymeraseX |
| POLG | 5428 | 2307441 | 1143 | E | G | 0.03 | PF00476 DNA_pol_A |
| POLG | 5428 | 2307440 | 1146 | R | C | 0.01 | PF00476 DNA_pol_A |
| POLG | 5428 | 2307442 | 1142 | R | W | 0.01 | PF00476 DNA_pol_A |
| POLI | 11201 | 3218778 | 71 | R | G | 0.01 | PF00817 IMS |
| POLI | 11201 | 3218787 | 535 | C | R | 0.01 | |
| POLI | 11201 | 3218786 | 507 | F | S | 0.01 | |
| POLL | 27343 | 3730477 | 438 | R | W | 0.13 | PF00966 DNA_polymeraseX |
| RFC2 | 5982 | 3135684 | 232 | A | V | 0.01 | PF00004 AAA |
| SOD2 | 6648 | 5746129 | 156 | R | W | 0.01 | PF02777 sodfe_C |
HUGO symbol.
LocusLink identifier (http://www.ncbi.nlm.nih.gov/LocusLink).
Reference SNP cluster identifier (http://www.ncbi.nlm.nih.gov/SNP).
Amino acid position in coding sequence.
Amino acid substitution of higher frequency allele in the PDR.
Amino acid substitution of lower frequency allele in the PDR.
Estimated frequency of minor allele in 180 chromosomes.
Protein family (http://www.sanger.ac.uk/Software/Pfam).
Of the 57 cSNPs predicted to be intolerant, seven are reported to be associated with a known phenotype (three of which intersect with the set of seven common predicted intolerant SNPs mentioned above). BRCA1 Q356R (MAF = 1%) is implicated in the breast cancers of a mother and two daughters of a Swiss family (Schoumacher et al. 2001); ARSA P82L (MAF = 1%) is associated with metachromatic leukodystrophy (Barth et al. 1995); CYP2C9 R144C (CYP2C9*2, MAF = 6%) is associated with low phenytoin metabolism in a Turkish population (Aynacioglu et al. 1999); MTHFR E429A (MAF = 27%) increased risk of neural tube defects in a Dutch study due to decreased folate metabolism in individuals who are compound heterozygotes with the A222V allele (van der Put et al. 1995). We also detected the A222V allele (MAF = 22%) in our resequencing, which is associated with increased plasma homocysteine levels and may be a risk factor in cardiovascular disease (Frosst et al. 1995). PMS1 G501R (MAF = 1%) is associated with nonpolyposis colon cancer in French families (Wang et al. 1999), and NAT2 R64Q (MAF = 2%) is associated with slow acetylation of carcinogenic arylamines and increased risk of bladder cancer in an African population (Bell et al. 1993).
To gauge the potential for functional consequences of the remaining 50 variants identified as “intolerant,” by both SIFT and Polyphen, we determined the number of the SIFT “intolerant” classifications for 32 candidate genes with known disease mutations. For these 32 genes, we queried all known mutations (n = 545) from the Human Gene Mutation Database (Stenson et al. 2003) and analyzed them by using SIFT. In this set, 369 (68%) mutations were classified as “intolerant” versus 176 (32%) as “tolerant” (Supplemental Table 4). Of the 140 known polymorphisms in these genes, derived from reports to the GeneSNPs database, 39 (28%) were identified as “intolerant.” These results are similar to frequencies reported in the validation of SIFT (Ng and Henikoff 2003), which confirms the sensitivity of this analysis and suggests that the 50 intolerant cSNPs unassociated with phenotypes in Table 1 represent a pool of candidate loci that should be interrogated in association studies.
We also identified eight variations predicted to truncate or alter protein translation (Table 2). SMUG1, HGF, RAD23A, and ERCC4 had nonsense SNPs that predict truncation at positions 136, 1156, 140, and 2169, respectively, in the polypeptides. RAG1, MSH6, and MGST2 had insertion/deletion polymorphisms that predict an altered reading frame and premature termination of translation. We identified a one-base insertion/deletion in codon 461 of RAG1. MSH6 has a 4-bp insertion/deletion in codon 4159 that predicts a nonsynonymous K4159D substitution and truncation of the last two amino acids. MGST2 has a one-base insertion/deletion in codon 352. GTF2H3 contains a SNP that abolishes the start codon. With the exception of SMUG1, all of these polymorphisms were found in the heterozygous state in a single individual for an allele frequency of <1% in the PDR90. The SMUG1 nonsense cSNP was observed in two heterozygotes in the PDR 90 panel (MAF = 1%).
Table 2.
Nonsynonymous cSNPs Predicted to Alter Translation
| Symbola | Substitutionb | Codonc | Frequencyd |
|---|---|---|---|
| SMUG 1 | Non-sense | 136 | 1% |
| HGF | Non-sense | 1156 | <1% |
| RAD23A | Non-sense | 140 | <1% |
| ERCC4 | Non-sense | 2169 | <1% |
| RAG1 | Insertion/deletion | <1% | |
| MSH6 | Insertion/deletion | <1% | |
| MGST2 | Insertion/deletion | <1% | |
| GTF2H3 | Abolished start condon | 1 | <1% |
HUGO symbol.
Consequence of substitution.
Codon in which cSNP occurs.
Minor alele frequency in the PDR 90.
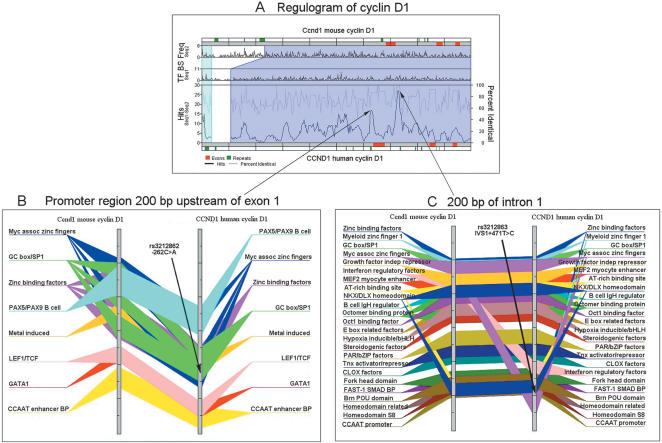
Many approaches are emerging to identify functional sites in noncoding regions. Trafac (Transcription Factor binding site Comparison) is a Web-accessible tool for identifying transcription regulatory regions by using a comparative sequence analysis approach (Jegga et al. 2002). Trafac generates a graphical output from BLASTZ alignments by comparing sequences of human and mouse orthologs or other sequences of interest. Potential transcription factor binding sites are identified as conserved blocks in the two sequences that are compared. An example of the Trafac output for CCND1 is shown in Figure 3. SNPs in conserved consensus transcription factor binding sites, such as the variation rs3212862 in a SP1 consensus site in the 5′ flanking sequence (Fig. 3B) and rs3212863 in the first intron (Fig. 3C), could adversely affect regulation of gene expression. The Trafac Web site is at http://genometrafac.cchmc.org/.
Figure 3.
Conserved noncoding regions identified by Trafac for Cyclin D (CCND1). (A) The regulogram depicts shared cis-elements between human and mouse sequences in the context of their sequence similarity. By identifying conserved mouse-human regions with consensus cis-regulatory elements and mapping noncoding SNPs, Trafac can be used to predict the potential adverse affects of polymorphisms on the regulation of gene and expression. Mouse and human sequences are represented as horizontal bars at the top and bottom of the upper pane. The red-colored segments on these bars represent exons 1 through 3. The green-colored bars represent repeat elements. The frequencies of individual binding sites occurring in each of the sequences separately are shown as two running graphs in the top half of the pane. The percentage of sequence similarity, as determined by the BLASTZ algorithm, and the number of transcription factor-binding sites (TF BS) is represented as two separate line graphs in the lower pane. Two cis-element dense regions within the highly conserved promoter and first intronic regions are depicted as two peaks with high hit (shared cis-elements) count (indicated by the arrows). The promoter (B) and first intronic regions (C) of human and mouse CCND1 reveal a strong conservation of consensus TF-binding sites in relatively the same order of occurrence. The two gray vertical bars represent the CCND1 orthologs. The TF BS occurring in both the genes are highlighted as various colored bars drawn across the two genes. The SNPs identified in the promoter (rs3212862) and first intron (rs3212863) are indicated.
Site Selection for Indirect Association Studies
Indirect association studies rely on linkage disequilibrium between genetic markers to measure the association between the SNP genotyped, as well as the SNPs in linkage disequilibrium with the assayed site and the disease phenotype (Collins et al. 1997). The number of sites required for genotyping in this study design depends on the strength and extent of linkage disequilibrium. For regions with strong linkage disequilibrium or few haplotypes only a few sites are required to represent or “tag” the region. However, if the genomic region contains many haplotypes indicating low levels of linkage disequilibrium, many more sites will be required for an association study of sufficient power. Given the variability of linkage disequilibrium described thus far across the human genome (Wall and Pritchard 2003), it is imperative to characterize the amount of site correlation a genomic region of interest contains if sites are to be rationally selected for genotyping in an association study.
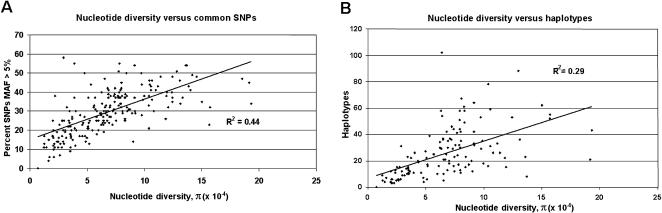
Therefore, to provide insight into the process of site selection to facilitate indirect association studies for these 213 candidate environmental response genes, we examined the extent of correlation between nucleotide diversity with two metrics of genomic variation: common SNPs (MAF > 5%; Fig. 4A) and haplotypes (Fig. 4B). Intragenic nucleotide diversity was modestly correlated (r2 = 0.44, P < 0.001) to the frequency of common SNPs (Fig. 4A). This was not unexpected because this measure of nucleotide diversity is sensitive to allele frequency. However, only 29% of the variability in the number of haplotypes per gene is associated with variability in nucleotide diversity per gene (r2 = 0.29; P < 0.001; Fig. 4B). Indeed, considerable gene-to-gene variation was observed in the number of haplotypes per gene, ranging from a low of three (FEN1) to a high of 102 (CCND2). Overall, the mean number of haplotypes per gene for the EGP was 26, which is lower than previous estimates from a set of genes related to inflammation, blood pressure regulation, and lipid metabolism (Crawford et al. 2004b). Both selection and recombination influence the shape of haplotype diversity, which could explain differences between genes and between gene data sets (Jorde et al. 2000). Also, it is important to keep in mind that the stratified nature of the PDR panel can have effects on both the haplotype diversity and patterns of linkage disequilibrium (LD). Nevertheless, given that the PDR broadly represents a United States population, these observations suggest that generalizations concerning haplotype diversity and the number of sites required to represent the haplotypes in a given candidate gene are problematic, and it is preferable to have some prior knowledge of these patterns before initiating a study.
Figure 4.
The relationship between nucleotide diversity and the number of common SNPs (A) or the number of inferred haplotypes (B) for 213 environmental response genes.
The gene-to-gene variability observed by the EGP in nucleotide diversity is also evident by site correlations or LD. Figure 5 illustrates the extremes observed in LD, as measured by the metric r2, across the genes involved in environmental responses. For genes with average or high LD, such as BNIP1 (Fig. 5A) and BRCA1 (Fig. 5C), respectively, few sites are required for genotyping in association studies. However, for genes with very weak LD, such as CCND2 (Fig. 5B), many more sites will be required for a genetic association study because very few sites within this gene are correlated. It is important to note that the extent of LD across a gene is independent of gene size. For example, LD extends across the 85-kb BRCA1, whereas fewer correlated sites are present in the smaller CCND2. For these genes with weak LD, attempts to choose sites with either LD-based (Carlson et al. 2004) or haplotype-based (Johnson et al. 2001) selection will require typing a larger fraction of the common sites in the candidate gene. Although the stratified nature of the PDR can produce artifactual LD, the patterns of LD described here represent the range of observed patterns within the EGP data set. Particularly for genes that exhibit strong LD (e.g., BRCA1), these patterns appear to be consistent among the ethnic subpopulations in the PDR, as few departures from Hardy-Weinberg equilibrium are observed.
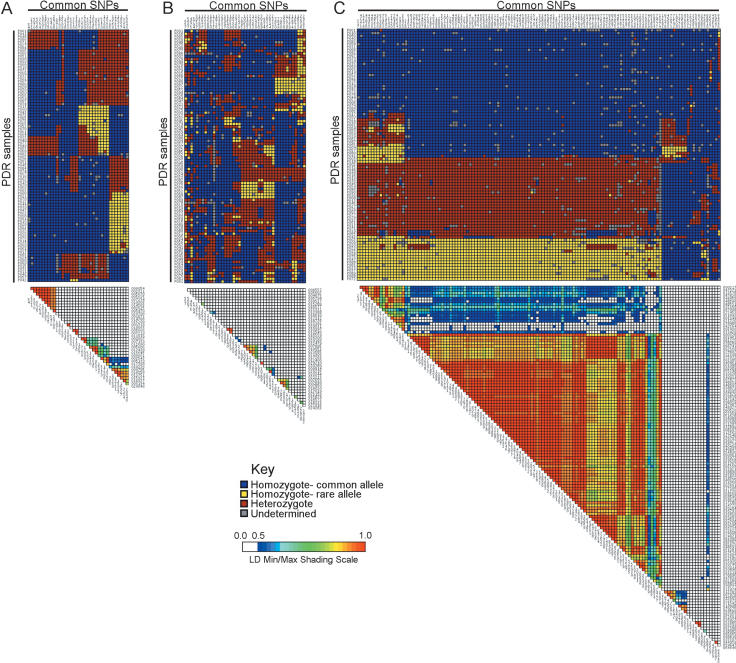
Figure 5.
Examples of linkage disequilibrium, as measured by r2, in candidate environmental response genes. (A) BNIP1 exhibits average LD, (B) CCND2 exhibits low LD, and (C) BRCA1 exhibits strong LD. The top portion of each graphic illustrates the visual genotypes for each gene, in which each column represents a site (blue indicates common homozygote; yellow, rare homozygote; red, heterozygote; and gray, missing data) and each row represents an individual from the PDR. The bottom portion of each graphic is the LD plot for each gene, measured by r2, and depicted on a rainbow scale (white indicates weak LD; red, strong LD).
DISCUSSION
This study presents one of the most comprehensive sets of genebased SNPs assembled, including both coding and noncoding SNPs, and provides an important view of the structure of sequence diversity across the human genome. Our analysis of ∼1% of the potential candidate genes located in the human genome reveals the range of gene-to-gene variation in overall nucleotide diversity, linkage disequilibrium, and number of haplotypes. As previously described for coding regions, there is wide-ranging gene-to-gene variation in coding region SNPs (Cargill et al. 1999; Halushka et al. 1999; Stephens et al. 2001). However, our report is one of the first to explore large amounts of noncoding genic sequence as well.
By extrapolating our findings from 213 candidate genes to the human genome, containing an estimated 24,000 to 35,000 total genes (Ewing and Green 2000; Lander et al. 2001), we estimate that the 1330 cSNPs identified in coding regions predict the presence of 150,000 to 219,000 cSNPs in the complete set of human genes (with a MAF >1%), similar to prior predictions (Kruglyak and Nickerson 2001). On average, 40% of the cSNPs are predicted to lead to amino acid substitutions. Therefore, we estimate that 60,000 to 87,600 amino acid-altering SNPs are present in the human genome.
By using SIFT and Polyphen to score potential functionally intolerant nonsynonymous cSNPs, we identified 57 SNPs predicted to alter protein function. Combined with the eight variations predicting altered polypeptide translation (four nonsense SNPs, three frameshifts, and one abolished start codon), the extrapolation of these observations predicts 7300 to 18,500 (assuming there are 24,000 to 35,000 genes) potentially deleterious SNPs in all human genes (with MAF > 1%). Of these 65 potential intolerant polymorphisms, only seven had a MAF >5%, suggesting a genic set of 790 to 1150 common deleterious SNPs. Although this estimate could potentially reflect relatively high conservation from a functional bias of these 213 genes, the gene-to-gene variation we observe in different measures of sequence diversity is consistent with previous observations of sets of genes encoding proteins involved in inflammation, lipid metabolism, and endocrine function (Cargill et al. 1999; Halushka et al. 1999; Stephens et al. 2001; Carlson et al. 2004; Crawford et al. 2004b). However, because similar constraints may exist in these other candidate gene sets, and no large-scale sequence surveys of random genes across the genome have been performed to date, it remains possible that this is a low estimate.
Informing Association Studies
That the haplotype structure and nucleotide diversity across the 213 genes we have studied are not well correlated suggests the design of candidate gene-based disease association studies will need to be tailored to the characteristics of the loci under consideration. Simple candidate gene association study designs using a uniform SNP density across all loci run the risk of either poorly characterizing high diversity loci, and/or over characterizing relatively simple loci, so investigators should use sequence diversity data when they are available. Rational site selection requires a complete view of the sequence diversity of the regions to be studied in order to accurately ascertain the potential association power of a given study design. Several obvious cSNP candidates were identified in this study; the 65 potentially functional variants are high priority candidates for direct association studies, although only a small fraction of this set is common in the population. According to the common disease/common variant hypothesis, the genetic risk factors underlying common diseases are likely common, modest-risk alleles in the human population (Chakravarti 1999; Altshuler et al. 2000; Reich et al. 2001). Of the seven common variants, three are listed in the HGM database as associated with phenotypes (CYP2C9 R144C, MTHFR A222V, and MTHFR E429A). The four unlisted common variants are POLL R438W, GSTZ1 E32K, GCKR P446L, and ERCC5 D1104H with allele frequencies of 13%, 30%, 33%, and 38% respectively. The POLL variant is in the pol X domain of the DNA polymerase. POLL, expressed highest in fetal liver and testis (Aoufouchi et al. 2000), is proposed to function in DNA repair in meiosis during spermatogenesis (Garcia-Diaz et al. 2000). The GSTZ1 variant (also called GST Z1B) is located in a β-strand of the protein and has little effect on the glutathione transferase activity for various substrates (Blackburn et al. 2001). The variant in the glucokinase regulatory protein, GCKR, may be a candidate SNP for maturity-onset diabetes of the young (MODY) due to the association of mutations in the glucokinase gene, GCK, in 20% of MODY in the United Kingdom (Thomson et al. 2003). The ERCC5 (also known as XPG) variant was reported as a polymorphism in a study of prostate cancer (Hyytinen et al. 1999) and is not associated with xeroderma pigmentosum.
To be explored further are the variants in the regulatory regions and conserved noncoding regions of these genes. Our observations of a general trend of lower nucleotide diversity in conserved noncoding regions identified by cross-species comparisons suggest these regions may be undergoing selection in human populations. Further refinement of these regions will become practical as more mammalian genomes are sequenced. The 767 variants in conserved noncoding regions have the potential to dys-regulate expression levels and alter target-cell specificities of their respective genes and contribute to the development or progression of environmental diseases (see Supplemental Table 3). Examination of these polymorphisms in the context of gene feature views such as the “regulograms” provided by tools like Trafac, as shown in Figure 3, will facilitate these studies by identifying candidate SNPs in consensus transcription factor binding site regions.
Our limited ability to a priori predict SNPs with functional consequences has led to the development of large-scale projects to discover and type common variation to identify regions with high and low linkage disequilibrium and haplotypes (The International HapMap Consortium 2003). With these data, indirect association studies can be designed to exploit the linkage disequilibrium across these regions to capture potential multisite associations using judicious site selection while maximizing the power of the study design.
Future of the EGP
The first phase of the EGP is completed (Kaiser 2003). In the next few years, the pace of EGP SNP discovery will continue to accelerate, and an additional 350 genes involved in cell signaling, cell structure, cell division, and metabolism will be studied. The next phase will also explore any biological relevance and functional significance of the 23,443 SNPs identified in phase 1 and interrogate these variants in molecular epidemiology studies of environmentally induced disease. Already we are seeing rapid advances as our data are mined as candidate causal polymorphisms in association studies (Ladiges et al. 2004). The final stage of the EGP will be realized when these studies translate into the decision-making that creates better environmental health policy and health monitoring practices with sensitivity and specificity for at-risk individuals.
METHODS
Candidate Environmental Response Genes
The targeted candidate genes for the EGP encode well-characterized groups of interacting proteins involved in pathways for DNA repair, cell cycle control, drug metabolism, and apoptosis. The candidate genes were selected by soliciting recommendations from investigators studying toxicogenomics and environmental susceptibility (Olden and Wilson 2000). Targeted candidate genes are listed on the EGP Web site (http://www.niehs.nih.gov/envgenom/home.htm) and on the GeneSNPs Web site (http://www.genome.utah.edu/genesnps).
SNP Discovery
SNPs were identified by amplifying the candidate genes from the 90 genomic DNA samples of the PDR and sequencing the amplification products. Primers were designed to amplify overlapping 800 to 1000 bp genomic fragments by using custom software (PCR-overlap, http://droog.mbt.washington.edu/PCR-Overlap.html). All exons, genomic sequences conserved in mouse, and >1 kb of 5′ and 3′ flanking regions were targeted. For genes <30 kb, the entire genomic sequence for the candidate was targeted for resequencing. For genes >30 kb, all coding and conserved noncoding sequence, as well as 20% of the remaining intronic sequences, were targeted. When feasible the EGP targeted alternatively transcribed exons (e.g., both exons 1 were resequenced for the unusual gene CDKN2A, which encodes two distinct cell cycle proteins, p14ARF and p16INK4A, that are alternatively transcribed using the different first exons and translated in different reading frames). Routinely, the EGP targets the longest genomic region corresponding to an alternative transcript or to the transcript that contains the most exons. Amplicons were designed to provide ∼20% redundancy of sequence coverage. This served to validate the primer sequence in independent experiments and rule out the possibility of allele-specific PCR amplification. Primer sequences included a universal M13 5′-18 base sequence for priming of sequencing reactions. All primer sequences were compared to the whole genome assembly to verify uniqueness against pseudogenes and gene families. Primer sequences are available at http://egp.gs.washington.edu/finished_genes.html.
The study population consisted of 90 DNA samples obtained from the PDR (Collins et al. 1998), a publicly available, anonymous collection of individuals that is representative of the ethnic diversity of the United States population (Coriell Institute; http://locus.umdnj.edu/nigms/products/pdr.html). The panel includes 24 European, 24 Asian, 24 African, 12 Hispanic, and six Native American samples. PCR amplifications were performed in 96-well plates in a volume of 7 μL comprising 0.2 μL each of 7 μM forward and reverse primers, 2.8 μL DNA (5 ng/μL), and 0.4 μL Elongase Enzyme (Invitrogen) per well. Reactions were incubated for 30 cycles by using optimized annealing temperatures in MJ Tetrad thermocyclers (PTC 225) with 96-well α units. Following evaluation by 1% agarose gel electrophoresis, reactions were diluted four- to sixfold in ddH2O. Dilution of the products eliminated the need for any purification of the products prior to sequencing. Sequencing reactions were performed in MJ Tetrad PTC 225 thermocyclers in 384-well format by using 5% BDT v3.1 sequencing chemistry (ABI). Reactions were precipitated in ethanol with CleanSeq magnetic beads (Agencourt). Packard Minitrack and Multiprobe robots were used to automate liquid handling in the setup of PCR and sequencing reactions.
Reaction products were air dried and diluted to 30 μL with ddH2O. Chromatograms were generated from reaction products on Applied Biosystems ABI 3700 or ABI 3730 capillary sequencers. Data flow was tracked by using a custom-designed LIMS system.
All chromatograms were base-called by using Phred, assembled into contigs by using Phrap, and scanned for SNPs with Polyphred, version 4.1 (Nickerson et al. 1997). Data quality was monitored and assessed at multiple production checkpoints and in a number of ways. Each chromatogram was trimmed to remove low-quality sequence (Phred score <25), resulting in analyzed reads averaging >450 bp with an average Phred quality of 40. We obtained second-strand confirmation from a different sequencing primer at 66% of all polymorphic sites and third-strand confirmation at 33% of all polymorphic sites. We observed all three possible genotypes (heterozygotes and homozygotes for each allele) for ∼38% of common polymorphic sites, with an average Phred quality >45 (1/50,000 probability of being incorrectly assigned). The average flanking sequence quality associated with polymorphic sites (±5 bp on each side of the polymorphic site) was >40. All sequence contigs were assembled and variant sites visually inspected by sequence analysts by using Consed. Variations with this level of accuracy were deposited into a custom postgres database and submitted to dbSNP.
Identification of Potential Functional Variants
SIFT (Sorting Intolerant From Tolerant; Ng and Henikoff 2003) and PolyPhen (Ramensky et al. 2002) were used to predict the effect of a nonsynonymous cSNP on protein function. The SIFT score is the normalized probability of a substitution at each position in the multiple alignment, based on the amino acids appearing at each position in the alignment of all orthologous polypeptides. The threshold for all columns of the position-specific scoring matrix, derived by using the query sequence against the consensus sequence seed obtained by PSI-BLAST, was 0.05. Substitutions at each position with normalized probabilities <0.05 were predicted to be intolerant. All cSNPs were mapped onto known protein domains, where available, by using the Polydoms server (http://polydoms.cchmc.org) and Pfam (http://www.sanger.ac.uk/Software/Pfam/) server (Bateman et al. 2004).
Identification of Conserved Noncoding Sequence
BLAST analysis was performed with repeat-masked (http://ftp.genome.washington.edu/RM/RepeatMasker.html; A.F.A. Smit and P. Green unpubl.) genomic sequence queries, using standard BLAST parameters, against the wgs.00 and wgs.01 BLAST databases, downloaded December 15, 2003, from the National Center for Biotechnology Information ftp site. The wgs databases contain contig assemblies from the whole-genome shotgun genome projects, including mouse, rat, and dog. Potential orthologous contigs from mouse, rat, and dog hits were searched against the July 2003 human reference sequence (UCSC version hg16), and contigs were included for analysis if they represented a reciprocal best hit to the initial human genomic query. The contigs were formatted for BLAST analysis and searched with human genomic queries by using the local blastall command with the following arguments: -F F (no low complexity filter), -E 2 (gap-extension penalty of one, default is two), and -m 3 (alignment view option, flat query-anchored, show identities). Composite ranges of genomic query sequence corresponding to mouse, rat, or dog HSPs (high scoring segment pairs) were parsed from the BLAST output. These ranges were then filtered based on the following criteria: size >50 bp, not contiguous (±8 bp to encompass splice sites) with known exons (GeneSNPs database), known mRNA ranges or GenScan predicted exons (UCSC Genome Browser), and the presence of EGP sequence coverage (any part of the range not within EGP resequencing was excluded).
Identification of Putative cis-Regulatory Regions
The Trafac server (Jegga et al. 2002) was used to identify putative cis-acting regulatory regions in EGP genes. The human genomic sequence for each of the genes was downloaded from the Gene SNPs (http://www.genome.utah.edu/genesnps/) database and the orthologous mouse sequence was obtained from the UCSC mouse database. The repeat-masked orthologous genomic sequences were aligned by using Advanced PipMaker (BLASTZ) with the chaining option (http://bio.cse.psu.edu/). MatInspector Professional version 4.3 (http://www.genomatix.de/; Quandt et al. 1995) was used to locate putative transcription factor binding sites using the TRANSFAC database (http://transfac.gbf.de; Wingender et al. 2000). The Trafac server integrates results from these applications and generates graphical outputs, the Regulogram and Trafacgram, examples of which are shown in Figure 3. Results for all EGP genes can be accessed at http://genometrafac.cchmc.org.
Haplotype Analysis
Haplotypes were inferred for 128 genes in which >75% of the gene was resequenced for SNP discovery by the statistical software package PHASE, version 2.0 (Stephens et al. 2001; Stephens and Donnelly 2003), which allows for missing genotype data, accommodates a large number of sites per gene, and accounts for recombination. Haplotypes were inferred by using the default settings of PHASE from genotype data using biallelic polymorphisms with a MAF >5% present in the PDR. To address the possibility that unknown population structure may have influenced the haplotype counts reported here, we examined haplotype estimates for three genes resequenced for both the EGP and the Program for Genomic Applications (PGA; Crawford et al. 2004b): TGFB3, LTA, and LTB. The PGA project resequences genes involved in inflammation, lipid metabolism, and blood pressure regulation in 23 European Americans and 24 African Americans. We inferred haplotypes for these three genes resequenced for both the EGP and PGA with PHASE, version 2.0, using a MAF of >5%. For the PGA data, we inferred haplotypes for the African American sample, the European American sample, and the combined sample separately. We found that the inferred haplotypes counts for the PGA combined sample were similar to the EGP sample, respectively, for all three genes: 38 versus 39 (TGFB3), 15 versus 16 (LTA), and seven versus five (LTB). Further examination of these genes demonstrates that the PGA combined sample for the gene TGFB3 has significant population structure (African Americans with 34 haplotypes and European Americans with nine haplotypes) and may have a hotspot of recombination based on the African American data (Crawford et al. 2004a). However, neither population structure nor frequent recombination seems to artificially inflate the estimated haplotype counts in the combined sample. Thus, the estimated haplotypes counts for the EGP sample are expected to approximate the true counts of the combined populations represented by the PDR90.
Acknowledgments
We acknowledge the talented technical and analytical staff that produced and reviewed these data: Moon Wook-Chung, Monica A. Montoya, Christine Vo, Laura A. Witrak, Annie M. Sherwood, Mike Daniels, Amy N. Olson, Brent J. Leithauser, Tasha K. Downing, Becky Borrayo, J. Tucker Jackson, Christopher Baier, R. Luke Daniels, Christa L. Poel, J. Kristofer Sherwood, Dan Nguyen, Wendy S. Schackwitz, Peggy D. Robertson, Sally W. Chambers, Ann C. Braun, Katy E. Miyamoto, Jonathan Alder, Diane Dunn, M. Hadi Islam, and Allan Tingey. We thank Drs. Samuel Wilson and Elizabeth Maull for their critical review of this manuscript and Eric Torskey for his assistance in its preparation. This work supported by National Institute of Environmental Health Sciences grants N01-ES-15478 (D.A.N.), N01-ES-35501 (R.B.W.), and U01-ES-11038 (B.J.A.).
Footnotes
Supplemental material is available online at www.genome.org. All sequence data from this study have been submitted to GenBank and are available from our Web site at http://egp.gs.washington.edu and at other sites listed herein.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2730004. Article published online ahead of print in September 2004.
References
- Alonso, J., Garcia-Miguel, P., Abelairas, J., Mendiola, M., Sarret, E., Vendrell, M.T., Navajas, A., and Pestana, A. 2001. Spectrum of germline RB1 gene mutations in Spanish retinoblastoma patients: Phenotypic and molecular epidemiological implications. Hum. Mutat. 17: 412-422. [DOI] [PubMed] [Google Scholar]
- Altshuler, D., Hirschhorn, J.N., Klannemark, M., Lindgren, C.M., Vohl, M.C., Nemesh, J., Lane, C.R., Schaffner, S.F., Bolk, S., Brewer, C., et al. 2000. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 26: 76-80. [DOI] [PubMed] [Google Scholar]
- Aoufouchi, S., Flatter, E., Dahan, A., Faili, A., Bertocci, B., Storck, S., Delbos, F., Cocea, L., Gupta, N., Weill, J.C., et al. 2000. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 28: 3684-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aynacioglu, A.S., Brockmoller, J., Bauer, S., Sachse, C., Guzelbey, P., Ongen, Z., Nacak, M., and Roots, I. 1999. Frequency of cytochrome P450 CYP2C9 variants in a Turkish population and functional relevance for phenytoin. Br. J. Clin. Pharmacol. 48: 409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, M.L., Fensom, A., and Harris, A. 1995. Identification of seven novel mutations associated with metachromatic leukodystrophy. Hum. Mutat. 6: 170-176. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, D.A., Taylor, J.A., Butler, M.A., Stephens, E.A., Wiest, J., Brubaker, L.H., Kadlubar, F.F., and Lucier, G.R. 1993. Genotype/phenotype discordance for human arylamine N-acetyltransferase (NAT2) reveals a new slow-acetylator allele common in African-Americans. Carcinogenesis 14: 1689-1692. [DOI] [PubMed] [Google Scholar]
- Blackburn, A.C., Coggan, M., Tzeng, H.F., Lantum, H., Polekhina, G., Parker, M.W., Anders, M.W., and Board, P.G. 2001. GSTZ1d: A new allele of glutathione transferase ζ and maleylacetoacetate isomerase. Pharmacogenetics 11: 671-678. [DOI] [PubMed] [Google Scholar]
- Botstein, D. and Risch, N. 2003. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 33(Suppl): 228-237. [DOI] [PubMed] [Google Scholar]
- Buchholz, T.A., Weil, M.M., Ashorn, C.L., Strom, E.A., Sigurdson, A., Bondy, M., Chakraborty, R., Cox, J.D., McNeese, M.D., and Story, M.D. 2004. A Ser49Cys variant in the ataxia telangiectasia, mutated, gene that is more common in patients with breast carcinoma compared with population controls. Cancer 100: 1345-1351. [DOI] [PubMed] [Google Scholar]
- Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231-238. [DOI] [PubMed] [Google Scholar]
- Carlson, C.S., Eberle, M.A., Rieder, M.J., Yi, Q., Kruglyak, L., and Nickerson, D.A. 2004. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74: 106-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti, A. 1999. Population genetics: Making sense out of sequence. Nat. Genet. 21: 56-60. [DOI] [PubMed] [Google Scholar]
- Collins, F.S., Guyer, M.S., and Charkravarti, A. 1997. Variations on a theme: Cataloging human DNA sequence variation. Science 278: 1580-1581. [DOI] [PubMed] [Google Scholar]
- Collins, F.S., Brooks, L.D., and Chakravarti, A. 1998. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 8: 1229-1231. [DOI] [PubMed] [Google Scholar]
- Crawford, D.C., Bhangale, T., Li, N., Hellenthal, G., Rieder, M.J., Nickerson, D.A., and Stephens, M. 2004a. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36: 700-706. [DOI] [PubMed] [Google Scholar]
- Crawford, D.C., Carlson, C.S., Rieder, M.J., Carrington, D.P., Yi, Q., Smith, J.D., Eberle, M.A., Kruglyak, L., and Nickerson, D.A. 2004b. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am. J. Hum. Genet. 74: 610-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist, A., Hammond, J.B., Crane, R.K., Dunphy, J.V., and Littman, A. 1963. Intestinal lactase deficiency and lactose intolerance in adults: Preliminary report. Gastroenterology 45: 488-491. [PubMed] [Google Scholar]
- Doll, R. 1975. Pott and the path to prevention. Arch. Geschwulstforsch. 45: 521-531. [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 2000. Analysis of expressed sequence tags indicates 35,000 human genes. Nat. Genet. 25: 232-234. [DOI] [PubMed] [Google Scholar]
- Frosst, P., Blom, H.J., Milos, R., Goyette, P., Sheppard, C.A., Matthews, R.G., Boers, G.J., den Heijer, M., Kluijtmans, L.A., van den Heuvel, L.P., et al. 1995. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 10: 111-113. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz, M., Dominguez, O., Lopez-Fernandez, L.A., de Lera, L.T., Saniger, M.L., Ruiz, J.F., Parraga, M., Garcia-Ortiz, M.J., Kirchhoff, T., del Mazo, J., et al. 2000. DNA polymerase λ (Pol λ), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 301: 851-867. [DOI] [PubMed] [Google Scholar]
- Haemmerli, U.P., Kistler, H., Ammann, T., Marthaler, T., Semenza, G., Auricchio, S., and Prader, A. 1965. Acquired milk intolerance in the adult caused by lactose malabsorption due to a selective deficiency of intestinal lactase activity. Am. J. Med. 38: 7-30. [DOI] [PubMed] [Google Scholar]
- Halushka, M.K., Fan, J.B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239-247. [DOI] [PubMed] [Google Scholar]
- Hutchison, D.C., Cook, P.J., and Barter, C.E. 1970. Pulmonary emphysema and α1-antitrypsin deficiency. Clin. Sci. 38: 19P. [DOI] [PubMed] [Google Scholar]
- Hyytinen, E.R., Frierson Jr., H.F., Sipe, T.W., Li, C.L., Degeorges, A., Sikes, R.A., Chung, L.W., and Dong, J.T. 1999. Loss of heterozygosity and lack of mutations of the XPG/ERCC5 DNA repair gene at 13q33 in prostate cancer. Prostate 41: 190-195. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium 2003. The international HapMap project. Nature 426: 789-796. [DOI] [PubMed] [Google Scholar]
- Jegga, A.G., Sherwood, S.P., Carman, J.W., Pinski, A.T., Phillips, J.L., Pestian, J.P., and Aronow, B.J. 2002. Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res. 12: 1408-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, G.C., Esposito, L., Barratt, B.J., Smith, A.N., Heward, J., Di Genova, G., Ueda, H., Cordell, H.J., Eaves, I.A., Dudbridge, F., et al. 2001. Haplotype tagging for the identification of common disease genes. Nat. Genet. 29: 233-237. [DOI] [PubMed] [Google Scholar]
- Jorde, L.B., Watkins, W.S., Kere, J., Nyman, D., and Eriksson, A.W. 2000. Gene mapping in isolated populations: New roles for old friends? Hum. Hered. 50: 57-65. [DOI] [PubMed] [Google Scholar]
- Kaiser, J. 2003. Tying Genetics to the risk of environmental diseases. Science 300: 563. [DOI] [PubMed] [Google Scholar]
- Klotz, A.P. 1964. Intestinal lactase deficiency and diarrhea in adults. Am. J. Dig. Dis. 10: 345-354. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L. and Nickerson, D.A. 2001. Variation is the spice of life. Nat. Genet. 27: 234-236. [DOI] [PubMed] [Google Scholar]
- Ladiges, W., Kemp, C., Packenham, J., and Velazquez, J. 2004. Human gene variation: from SNPs to phenotypes. Mutat. Res. 545: 131-139. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Li, W.H. and Sadler, L.A. 1991. Low nucleotide diversity in man. Genetics 129: 513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman, J., Mittman, C., and Schneider, A.S. 1969. Screening for homozygous and heterozygous α1-antitrypsin deficiency: Protein electrophoresis on cellulose acetate membranes. JAMA 210: 2055-2060. [PubMed] [Google Scholar]
- Mathonnet, G., Krajinovic, M., Labuda, D., and Sinnett, D. 2003. Role of DNA mismatch repair genetic polymorphisms in the risk of childhood acute lymphoblastic leukaemia. Br. J. Haematol. 123: 45-48. [DOI] [PubMed] [Google Scholar]
- Motulsky, A.G. 1972. Hemolysis in glucose-6-phosphate dehydrogenase deficiency. Fed. Proc. 31: 1286-1292. [PubMed] [Google Scholar]
- Ng, P.C. and Henikoff, S. 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D.A., Tobe, V.O., and Taylor, S.L. 1997. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25: 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D.A., Taylor, S.L., Weiss, K.M., Clark, A.G., Hutchinson, R.G., Stengard, J., Salomaa, V., Vartiainen, E., Boerwinkle, E., and Sing, C.F. 1998. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat. Genet. 19: 233-240. [DOI] [PubMed] [Google Scholar]
- Olden, K. and Wilson, S. 2000. Environmental health and genomics: Visions and implications. Nat. Rev. Genet. 1: 149-153. [DOI] [PubMed] [Google Scholar]
- Quandt, K., Frech, K., Karas, H., Wingender, E., and Werner, T. 1995. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23: 4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky, V., Bork, P., and Sunyaev, S. 2002. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 30: 3894-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, D.E., Cargill, M., Bolk, S., Ireland, J., Sabeti, P.C., Richter, D.J., Lavery, T., Kouyoumjian, R., Farhadian, S.F., Ward, R., et al. 2001. Linkage disequilibrium in the human genome. Nature 411: 199-204. [DOI] [PubMed] [Google Scholar]
- Sachidanandam, R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928-933. [DOI] [PubMed] [Google Scholar]
- Schoumacher, F., Glaus, A., Mueller, H., Eppenberger, U., Bolliger, B., and Senn, H.J. 2001. BRCA1/2 mutations in Swiss patients with familial or early-onset breast and ovarian cancer. Swiss Med. Wkly. 131: 223-226. [DOI] [PubMed] [Google Scholar]
- Stenson, P.D., Ball, E.V., Mort, M., Phillips, A.D., Shiel, J.A., Thomas, N.S., Abeysinghe, S., Krawczak, M., and Cooper, D.N. 2003. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 21: 577-581. [DOI] [PubMed] [Google Scholar]
- Stephens, M. and Donnelly, P. 2003. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73: 1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, J.C., Schneider, J.A., Tanguay, D.A., Choi, J., Acharya, T., Stanley, S.E., Jiang, R., Messer, C.J., Chew, A., Han, J.H., et al. 2001. Haplotype variation and linkage disequilibrium in 313 human genes. Science 293: 489-493. [DOI] [PubMed] [Google Scholar]
- Sunyaev, S., Ramensky, V., Koch, I., Lathe III, W., Kondrashov, A.S., and Bork, P. 2001. Prediction of deleterious human alleles. Hum. Mol. Genet. 10: 591-597. [DOI] [PubMed] [Google Scholar]
- Thomson, K.L., Gloyn, A.L., Colclough, K., Batten, M., Allen, L.I., Beards, F., Hattersley, A.T., and Ellard, S. 2003. Identification of 21 novel glucokinase (GCK) mutations in UK and European Caucasians with maturity-onset diabetes of the young (MODY). Hum. Mutat. 22: 417. [DOI] [PubMed] [Google Scholar]
- van der Put, N.M., Steegers-Theunissen, R.P., Frosst, P., Trijbels, F.J., Eskes, T.K., van den Heuvel, L.P., Mariman, E.C., den Heyer, M., Rozen, R., and Blom, H.J. 1995. Mutated methylenetetrahydrofolate reductase as a risk factor for spina bifida. Lancet 346: 1070-1071. [DOI] [PubMed] [Google Scholar]
- Wall, J.D. and Pritchard, J.K. 2003. Haplotype blocks and linkage disequilibrium in the human genome. Nat. Rev. Genet. 4: 587-597. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Lasset, C., Desseigne, F., Saurin, J.C., Maugard, C., Navarro, C., Ruano, E., Descos, L., Trillet-Lenoir, V., Bosset, J.F., et al. 1999. Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer. Hum. Genet. 105: 79-85. [DOI] [PubMed] [Google Scholar]
- Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Pruss, M., Reuter, I., and Schacherer F. 2000. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 28: 316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://locus.umdnj.edu/nigms/products/pdr.html); Coriell Institute.
- http://www.niehs.nih.gov/envgenom/home.htm; Environmental Genome Project.
- http://www.genome.utah.edu/genesnps; GeneSNPs.
- http://www.genomatix.de; Genomatix.
- http://www.ncbi.nlm.nih.gov/LocusLink; NCBI LocusLink.
- http://www.ncbi.nlm.nih.gov/SNP; NCBI Single Nucleotide Polymorphism.
- http://egp.gs.washington.edu/finished_genes.html; NIEHS SNPs.
- http://droog.mbt.washington.edu/PCR-Overlap.html; PCR-Overlap.
- http://bio.cse.psu.edu; Penn State University Center for Comparative Genomics and Bioinformatics, Miller Lab (in collaboration with Ross Hardison).
- http://www.sanger.ac.uk/Software/Pfam/; Pfam.
- http://polydoms.cchmc.org; PolyDom.
- http://ftp.genome.washington.edu1/RM/RepeatMasker.html; RepeatMasker documentation.
- http://genometrafac.cchmc.org; Trafac.
- http://transfac.gbf.de; TRANSFAC.