FIG 4.
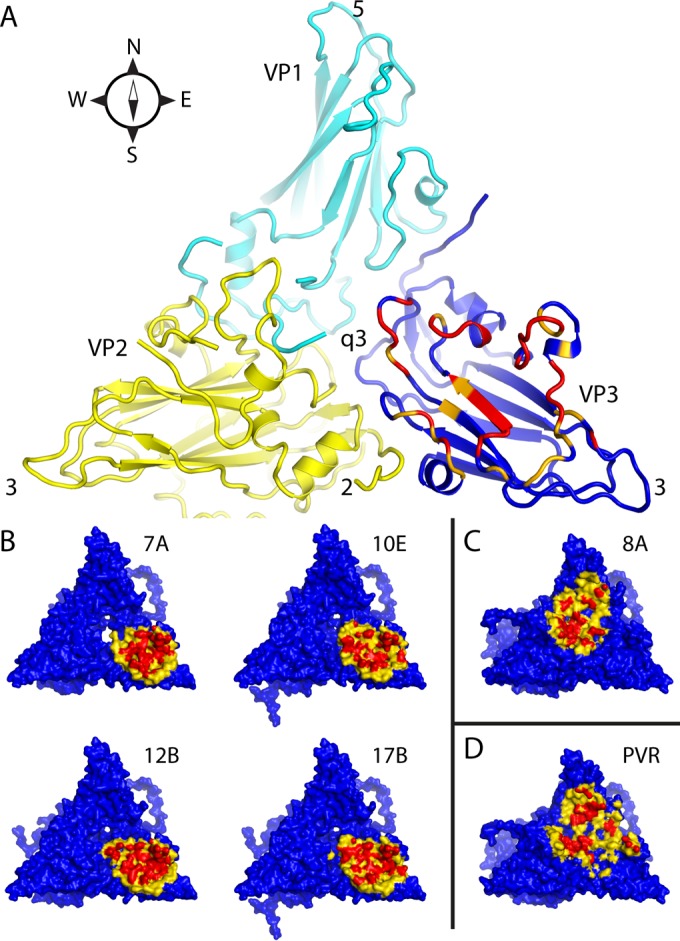
Intermolecular contact areas on the poliovirus surface (“footprints”) that are involved in binding to various proteins. (A) One copy each of VP1 (cyan), VP2 (yellow), and VP3 (mostly dark blue) occupy the “5-3-3 triangle,” which has one 5-fold and two 3-fold symmetry axes at its corners (labeled). Expansion of poliovirus opens holes at the 2-fold axis (labeled) and at the quasi-3-fold axis (labeled). The quasi-3-fold axis lies at the center of the 5-3-3 triangle and is the point at which these three capsid proteins join together in the native virus. By convention, compass directions (north, south, east, and west) indicate the relative positions along the virus surface. This ribbon representation shows the “footprint” of one copy of anti-C-VHH PVSP17B on expanded poliovirus. In VP3, which is the only capsid protein to contact the anti-C VHHs, residues that are responsible for binding are indicated in red (distances of <4.2 Å), and residues with contacts closer than 6.0 Å are orange or yellow. (B to D) Surface representations, color-coded by distance (red, 5 Å or closer; yellow, 5 to 9 Å), show that the binding footprints of the four anti-C VHHs on expanded poliovirus (B) are similar to one another but differ from the footprints (on native poliovirus) of poliovirus receptor (“PVR”) (D) and from the anti-N VHHs (C), of which PVSS8A (“8A”) is typical. Thus, Pvr and all five anti-N VHHs consistently bind to parts of VP1 and VP2 that are close to the quasi-3-fold axis. The surface representations make it apparent that the quasi-3-fold hole is visibly larger in the PVSS7A complex than it is in the other three anti-C VHH complexes.
