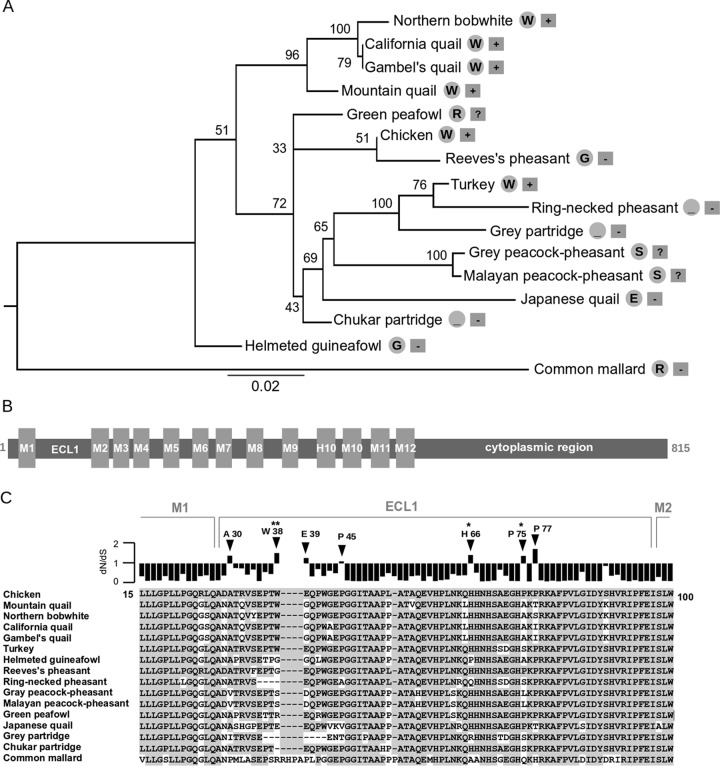
FIG 6.
Molecular evolution of NHE1 in galliform birds. (A) ML tree of NHE1 partial nucleotide sequences of galliform species with the common mallard as an outlier. Nonparametric bootstrap supports are given next to the corresponding nodes. For each species, the critical residue or deletion at position 38 relative to the chicken sequence, taken as a reference, is shown in a gray circle on the right. Information about the sensitivity of the species to ALV-J infection is shown in a gray rectangle on the far right (+, susceptible; −, resistant; ?, unknown). Bar, substitutions per nucleotide. (B) Domain structure of the NHE1 protein. Membrane regions (M1 to M12, H10), extracellular loop 1 (ECL1), and the C-terminal cytoplasmic domain are shown. Annotation is based on the human NHE1 ortholog. (C) Amino acid sequence alignment and positive-selection analysis of the ECL1 region. Residues identical to the chicken sequence are shown on a gray background. Ratios of nonsynonymous to synonymous substitution rates (dN/dS) for each alignment position are shown above the alignment. Residues with positive-selection signatures (dN/dS, >1) are indicated by arrowheads. Residues with a posterior probability for positive selection of >0.95 or >0.99 are marked with one or two asterisks, respectively. Positively selected sites were predicted by the REL method.