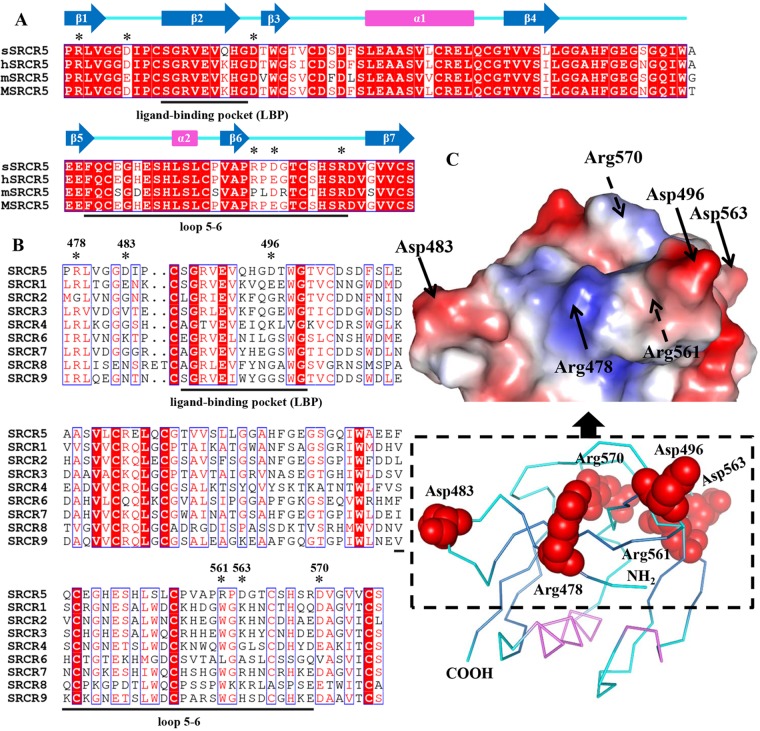
FIG 4.
Structure-based sequence alignment of the CD163 SRCR5 from different species and the nine SRCR domains of pCD163. (A) Sequence alignment of the CD163 SRCR5 from different species. The amino acid sequences of CD163 SRCR5 from pig (p; UniProt entry Q2VL90), human (h; UniProt entry Q86VB7), mouse (m; UniProt entry Q2VLH6), and monkey (M; UniProt entry Q2VLG4) are aligned. The amino acids within the so-called loop5-6 region and ligand-binding pocket (LBP) are underlined. Conserved charged residues in these two regions are colored red, and the mutated residues are marked by asterisks. The secondary structural elements of pCD163 SRCR5 are indicated above the aligned sequences. The blue arrows represent the β-strands from β1 to β7, and the violet rectangles represent two helices (α1 and α2). (B) Sequence alignment of the nine SRCR domains of pCD163 (UniProt entry Q2VL90). The amino acids within the so-called loop5-6 region and LBP are underlined. Conserved charged residues in these two regions are colored red, and the mutated residues are marked by asterisks. (C) Site-directed mutagenesis of pCD163 SRCR5. The ribbon diagram and the surface electrostatic potential map of pCD163 SRCR5 are shown with the mutated residues. The mutated residues are labeled as red spheres, and the N and C termini are labeled in the ribbon diagram. The mutated residues are also indicated with arrows in the surface electrostatic potential map, where the solid arrows indicate visible residues and the dashed arrows indicate ones not visible from the front view. The electrostatic potential is colored from −62 to +62 kiloteslas/charge.