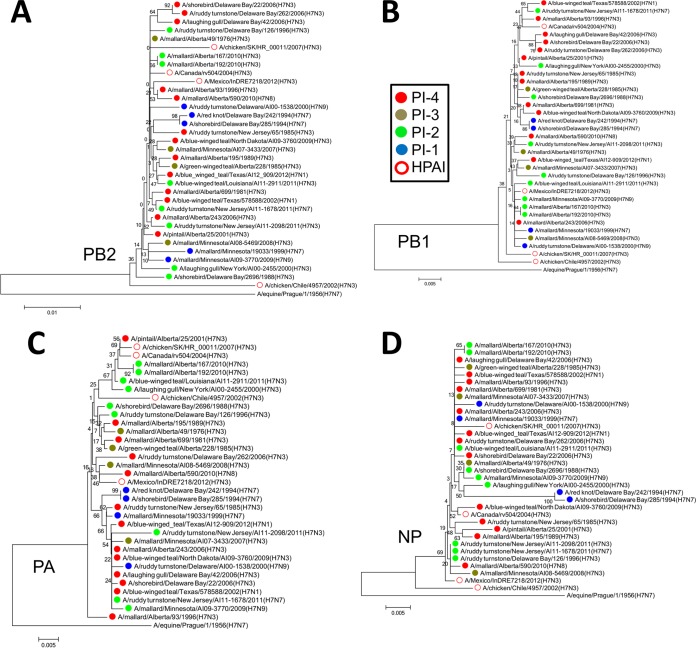
FIG 8.
Phylogenetic analysis revealed similarities between PB2, PB1, PA, and NPs based on the PI. (A) The PB2s of the HPAI viruses A/Mexico/InDRE7218/2012 (H7N3), A/Canada/rv504/2004 (H7N3), and A/chicken/SK/HR_00011/2007 (H7N3) clustered with each other and with those of PI-4 viruses. (B) PB1 of the HPAI virus A/Canada/rv504/2004 (H7N3) clustered with those of five PI-4 viruses and one PI-2 virus. (C) The PAs of the HPAI viruses A/Canada/rv504/2004 (H7N3) and A/chicken/SK/HR_00011/2007 (H7N3) clustered together, along with a those of PI-4 virus and several related PI-2 viruses. The PA of the HPAI virus A/chicken/Chile/4957/2002 (H7N3) also clustered with these viruses. The PA of A/Mexico/InDRE7218/2012 (H7N3) clustered with those of two PI-4 viruses and one PI-3 virus. (D) The NPs of A/Mexico/InDRE7218/2012 (H7N3) and A/Canada/rv504/2004 (H7N3) clustered with those of PI-4 viruses. The neighbor-joining trees were constructed with the full amino acid-coding sequence of each gene segment of the viruses using MEGA6 and rooted to sequences from A/equine/Prague/1/1956 (H7N7). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The evolutionary distances were computed using the Poisson correction method. The viruses were labeled according to their PI classifications.