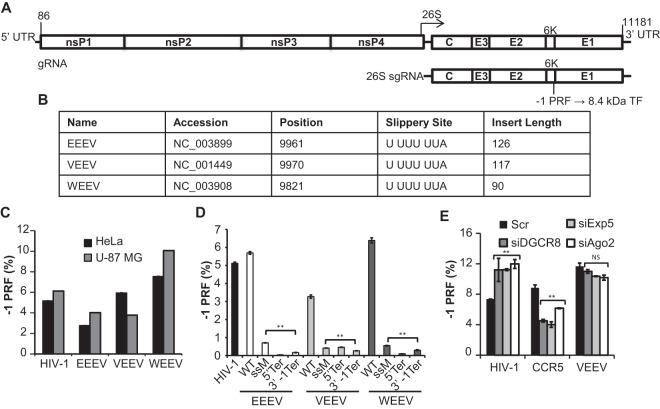
FIG 1.
Identification and monitoring of −1 PRF in the alphaviruses. (A) General schematic of the alphavirus genome and subgenomic transcript. The specific nucleotide numbers are given for the boundaries of the VEEV untranslated regions (UTRs). This family of viruses harbors −1 PRF signals toward the 3′ end of the 6K structural protein-encoding mRNA. The resulting frameshift product is an 8.4-kDa protein called the trans-frame (TF) protein. gRNA, genomic RNA; sgRNA, subgenomic RNA. (B) Accession numbers of the sequences from which the predicted −1 PRF signals were cloned, nucleotide positions of the first base of the predicted slippery sites, the slippery site sequences (the 0 frame is indicated by spaces), and the lengths of inserts (in numbers of nucleotides) initially tested for −1 PRF activities. (C) The predicted −1 PRF signals derived from EEEV, VEEV, and WEEV were cloned into the dual-luciferase reporter plasmid pJD175f, and their ability to promote efficient levels of frameshifting was measured in both HeLa cells and U-87 MG astrocyte cells. The −1 PRF signal from HIV-1 was employed as a positive control. (D) Site-directed mutagenesis was utilized to genetically validate the −1 PRF activities of the EEEV-, VEEV-, and WEEV-derived sequences in U-87 MG cells. WT, wild type; ssM, silent mutations of the slippery sites; 5′ Ter, 0-frame termination codons introduced upstream of the slippery sites; 3′ −1 Ter, termination codons inserted in the −1 reading frame 3′ of the virus-derived sequences. (E) miRNA processing or export was inhibited by siRNA knockdown of DGCR8 (siDGCR8), Exportin 5 (siExp5), or Argonaute 2 (siAgo2) in U-87 MG cells, and the rates of −1 PRF promoted by the indicated sequences were monitored. Control samples were transfected with siRNAs harboring scrambled (Scr) sequences. For assays of −1 PRF, a minimum of three or more biological replicates was performed in triplicate until statistical significance was achieved, as previously described (11). Bars represent standard errors of the means. **, P < 0.01; NS, not significant.