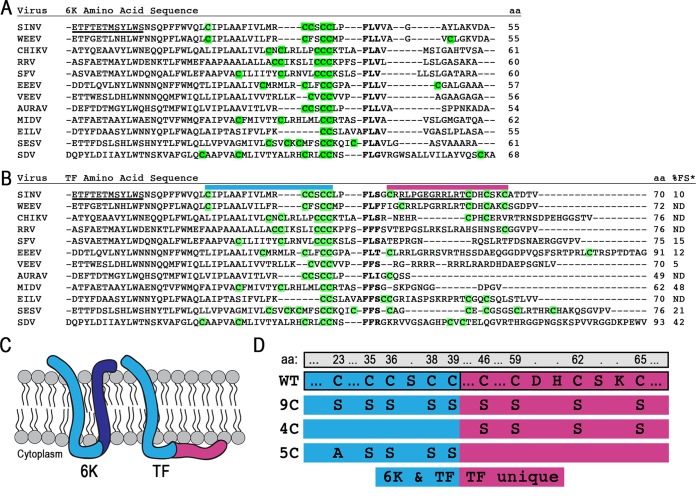
FIG 1.
The 6K and TF proteins and their Cys residues. (A and B) Protein sequences were aligned by using ClustalOmega and manually adjusted around the slip site (boldface type). All residues N terminal to the slip site are identical in 6K and TF, while residues C terminal to the slip site are unique to each protein. All Cys residues theoretically available for palmitoylation are highlighted in green. The protein length is given in amino acids. Underlined residues indicate N-terminal or C-terminal antibody epitopes. Above the sequence, colored bars correspond to regions magnified in panel D. The percent frameshifting rate (%FS), as measured by Chung et al. in a cell-based dual-luciferase assay (indicated by *), is shown at the far right of panel B (25). Abbreviations: SINV, Sindbis virus; WEEV, Western equine encephalitis virus; CHIKV, Chikungunya virus; RRV, Ross River virus; SFV, Semliki Forest virus; EEEV, Eastern equine encephalitis virus; VEEV, Venezuelan equine encephalitis virus; AURAV, Aura virus; MIDV, Middelburg virus; EILV, Eilat virus; SESV, Southern elephant seal virus; SDV, sleeping disease virus. (C) Schematic of the theoretical membrane topology of the 6K and TF proteins. The blue regions are sequences shared between both 6K and TF isoforms. The dark blue and red regions are unique to the C terminus of 6K and TF, respectively. (D) The mutations made for constructing the TF Cys mutants 9C, 4C, and 5C are demonstrated below the WT sequence. Amino acid (aa) numbering across the top is for WT Sindbis virus TF. The blue regions on the left are sequences shared between both 6K and TF isoforms. The red regions to the right are unique to the C terminus of TF.