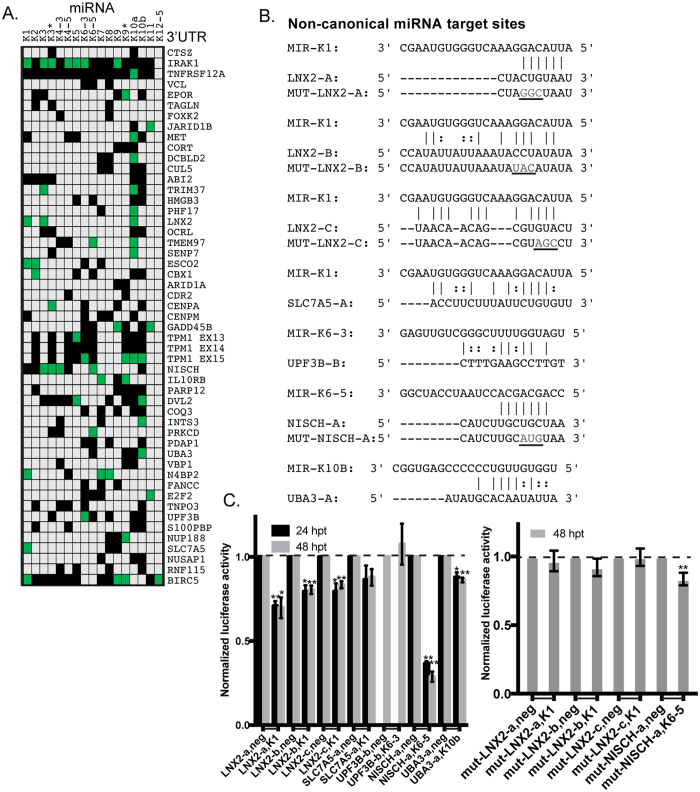
Figure 1. 3′UTR luciferase assays for predicted targets of KSHV-encoded miRNAs.
(A) 293 cells were transfected with 3′UTR luciferase reporter plasmids for the indicated genes along with KSHV miRNA mimics. Cells were harvested at 48 hpt and reporter activity was measured as described in Methods. The table highlights conditions where the luciferase reporter was repressed (p < 0.05) in green boxes. Gray boxes denote untested conditions and black boxes represent tested conditions where no significant repression was observed. (B) Potential non-canonical miRNA targets were identified using miRanda25 and PAR-CLIP sequences7. Mutated bases (“MUT”) are denoted in gray text and underlined. (C) Luciferase assays with the sites in (B) are shown. Activity with each miRNA was normalized to a negative control miRNA (Neg) with each reporter (denoted by joining horizontal line on x-axis). Error bars show standard deviation from three biological replicates. *Represents p < 0.05 and ** is p < 0.01.