Abstract
The structures of several powerful inhibitors of hydrolytic enzymes resemble that of the altered substrate in the transition state, except that a hydrogen atom replaces one substituent (typically the leaving group). To test the hypothesis that a water molecule might be present in the gap resulting from this replacement, we examined a transition-state analogue complex formed by Escherichia coli cytidine deaminase by Fourier transform ion cyclotron resonance MS in electrospray mode. Upon nebularization from aqueous solution under conditions (pH 5.6) where the enzyme is active, cytidine deaminase remains dimeric in the vapor phase. In the presence of inhibitor, the enzyme's exact mass can be used to infer the presence at each active site of zinc, 5-fluoro-3,4-dihydrouridine, and a single water molecule.
In the design of powerful enzyme inhibitors, the altered substrate in the transition state can be considered to represent an ideal ligand, for which the binding affinity of an enzyme has been optimized through natural selection. Because the properties of this short-lived species (t1/2 ≈ 10-13 s at 25°C) will never be matched exactly by those of any molecule with stable geometry, it is not surprising that even the most powerful inhibitors that have been designed on this basis are bound with an affinity (as judged by their Ki values as competitive inhibitors) that falls short of the ideal affinity that is estimated by comparing the second-order rate constant for the enzyme reaction (kcat/Km) with the first-order rate constant (knon) for the reaction in water in the absence of a catalyst (1, 2).
Some enzyme inhibitors do, nonetheless, capture much of the negative free energy of binding that would be expected of an ideal transition-state analogue, and the shortcomings of these unusually powerful inhibitors are of special interest. Several nucleoside deaminases and proteases, for example, are strongly inhibited by molecules in which a hydrogen atom replaces the leaving ammonia or amino group that would ordinarily be present in tetrahedral intermediates that approach the transition state in structure (Fig. 1). Such a truncated inhibitor would be expected to occupy much of the space that is normally occupied by the altered substrate in the transition state, except that a gap would presumably remain where the attacking or leaving group of the substrate had been replaced by a hydrogen atom. The appearance of that gap might result in the trapping of one or more water molecules, or the active site might collapse in such a way as to fill the gap. Either of these developments would be expected to be energetically unfavorable compared with the presumably optimal binding of the altered substrate in the transition state (3).
Fig. 1.
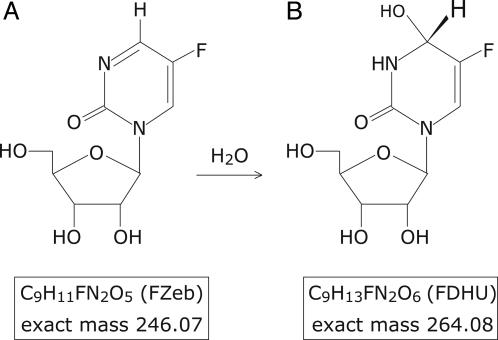
Chemical structure and exact mass of FZeb, also named 5-fluoro-zebularine (A), and its hydrated version, the transition-state analogue FDHU (B), which is obtained by the CDA-catalyzed 3,4-addition of water to FZeb.
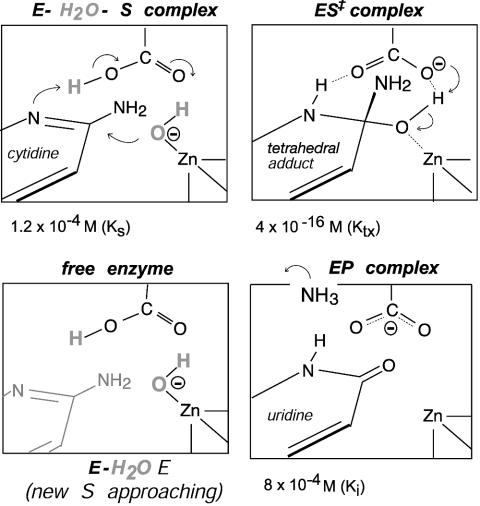
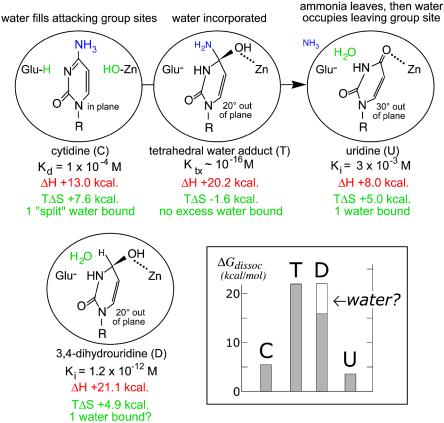
One enzyme for which this question has arisen is Escherichia coli cytidine deaminase (CDA), which consists of two symmetrical subunits (2 × 31,539 Da) with one active site per subunit (4). The active sites, which lie at the dimer interface, operate independently (5), and each is equipped with a zinc atom that is involved in the covalent addition of water to cytidine, followed by elimination of ammonia (Scheme 1) (6). The enzymatic and nonenzymatic reactions are both pH-independent, and each appears to proceed by a two-step mechanism that involves stepwise addition of zinc-coordinated hydroxide to form a tetrahedral intermediate, followed by rate-limiting elimination of NH3 (6). CDA is strongly inhibited by stable analogues resembling a transition state that is sp3-hybridized at C-4: 3,4-dihydrouridine (DHU) and its 5-fluorinated analogue, 5-fluoro-DHU (FDHU), which is formed by enzyme-catalyzed 3,4-addition of water to the corresponding 5-fluoropyrimidin-2-one ribonucleoside (FZeb) (Fig. 1). These analogues exhibit comparable affinities for CDA, surpassing the substrate or product in their negative free energies of binding by 10–11 kcal/mol. However, their affinities fall short of the affinity of the altered substrate in the transition state [estimated by comparing the rates of the enzymatic (kcat/Km) and nonenzymatic (knon) reactions] by 5–6 kcal/mol. In a recent investigation of the basis of this shortfall, the thermodynamic changes that accompany the binding of DHU by CDA were compared with those that accompany the binding of the distorted form of the substrate that is present in the transition state (7). The enthalpies of binding of DHU and the altered substrate in the transition state were found to differ by <1 kcal/mol, as would perhaps be expected whether similar polar forces of attraction were at work. However, the entropy of binding of this transition-state analogue (in which a hydrogen atom replaces the leaving amino group of cytidine) was found to be markedly less favorable (Scheme 2), and the magnitude of that difference was large enough to account for the shortcomings of the inhibitor as a ideal transition-state analogue.
Scheme 1.
Cycle of events during the enzymatic deamination of cytidine (4). Note that substrate water is present in “split” form on the free enzyme in solution.
Scheme 2.
Thermodynamic changes that accompany ligand binding by CDA (7).
To explain the less-than-ideal entropy of DHU, it was suggested that binding of this inhibitor might be accompanied by the unfavorable trapping of an extra water molecule within the active site, in the space vacated by the nitrogen atom of the missing 4-NH2 group (Scheme 2 Lower). During the enzyme-catalyzed reaction, the substrate's ribose substituent remains fixed in position within the active site, whereas the 4-carbon of the pyrimidine ring describes an arc with respect to the fulcrum of C-1′, away from the leaving group ammonia and toward the zinc-bound hydroxide ion (8). In the crystal structure of the enzyme–uridine complex, a water molecule was found to be present at the site considered to be occupied by the leaving group ammonia, in the normal reaction (8). The crystal structure of the enzyme's complex with FDHU (4) does not reveal the presence of such a water molecule, but does indicate the presence of a gap that may be large enough to accommodate a water molecule, at a position that would be sealed off from bulk solvent when the analogue was bound (7).
We considered it possible, at least in principle, that the presence of a water molecule in the inhibitory complex might be detected by accurate MS analysis of CDA in the presence and absence of a transition-state analogue inhibitor. In electrospray ionization MS analysis, proteins are ordinarily nebularized from acid solution to enhance the number of positive charges on the protein and reduce the value of m/z. That procedure tends to remove associated water molecules, revealing peaks that correspond to the mass of a “naked,” denatured form of the protein (9). Here, we describe the initial results of using Fourier transform ion cyclotron resonance (FTICR)-MS to analyze CDA, in the presence and absence of the inhibitor FZeb, under nondenaturating conditions (pH 5.6) where the protein retains activity in solution (5).
Materials and Methods
Recombinant CDA was expressed in E. coli and purified to homogeneity as described earlier (10, 11). The enzyme was dialyzed against ammonium acetate (5 × 10-3 M, pH 5.6) to produce a concentration of 5 × 10-6 M in monomer. For MS analysis, CDA was diluted with water to a final concentration of 1 × 10-6 M in monomer. Spectra of the acid-inactivated monomeric enzyme were obtained by analyzing CDA in the presence of 50% methanol and 2% acetic acid. Samples of the native enzyme in the presence and absence of inhibitor were acidified just before nebularization by addition of acetic acid, to produce a final concentration of 0.5%, without addition of other organic solvents. MS analyses were performed by using a hybrid Qq-FTICR mass spectrometer (Apex-Qq), equipped with either a 7- or 9.4-T actively shielded magnet (Bruker Daltonics), and an Apollo electrospray source. The capillary entrance voltage used to induce the electrospray ionization was between 3,900 and 4,200 V. Desolvation was carried out by using a countercurrent drying gas with a source temperature maintained at 150°C. The skimmer voltage applied (i.e., the potential difference between the skimmer and the capillary exit) varied between 164 and 181 V. Before transfer and detection, ion packets were accumulated in the collision cell of the system for a duration of 1–3 s. Samples were introduced by direct infusion with a 100-μl syringe (Harvard Apparatus) operated at a flow rate of 1 μl/min.
Results
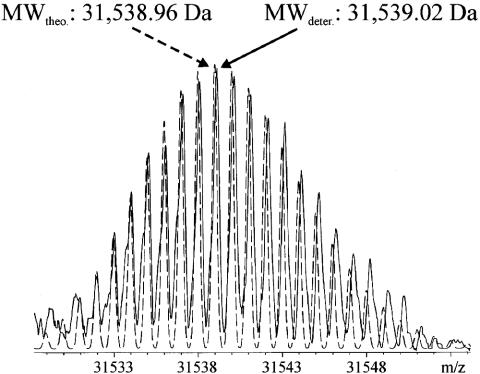
For accurate determination of the molecular mass of E. coli CDA, the protein was first analyzed under denaturing conditions, in a 50% methanol/2% acetic acid ratio, to disrupt the natural dimeric structure of the native enzyme in solution. By using a 9.4-T Bruker Qq-FTICR-MS, the mass spectrum showed isotopic resolution (Fig. 2), indicating 31,539 Da as the most abundant isotopic mass. That value is exactly equivalent to the mass expected from the elemental composition of the monomeric enzyme, with no zinc atoms and no associated water molecules. Subsequent analyses were performed by using a 7-T FTICR, because, for these experiments, the mass accuracy is easily sufficient for determination of the number of water molecules that are present in a protein.
Fig. 2.
FTICR-MS analysis of CDA under denaturing conditions by using a Bruker Apex Qq-FTICR equipped with a 9.4 T magnet. The deconvoluted spectrum obtained (solid line) is superimposed on the theoretical isotopic distribution of CDA (dashed line) derived from its elemental composition. The most abundant mass agrees with the theoretical mass within 2 ppm (0.13 Da). theo., theoretical; deter., experimentally determined; MW, molecular mass.
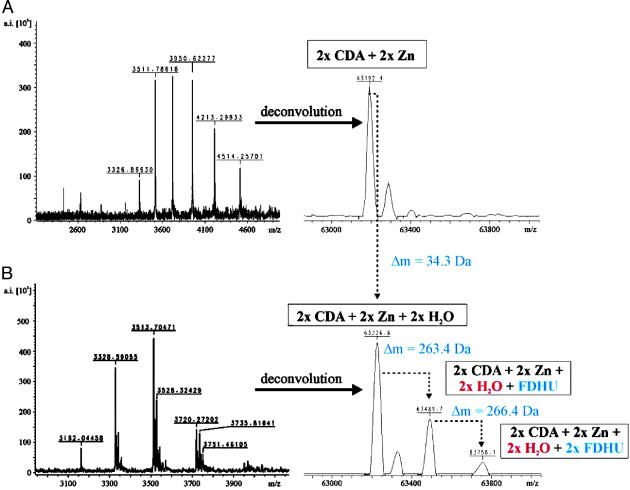
When we conducted this experiment by using enzyme dialyzed against ammonium acetate (0.005 M, pH 5.6), we observed one major ion series corresponding to the mass of the dimeric protein, containing two zinc atoms per dimer (Fig. 3A). We observed that methanol, even in small amounts, prevented dimer formation, but that acetic acid (0.5%), added just before nebularization, was necessary to obtain ion signals related to dimeric CDA, which contains one zinc atom at each of its active sites.
Fig. 3.
FTICR-MS analysis of CDA under nondenaturing conditions by using a Bruker 7-T Apex Qq-FTICR in the absence (A) and the presence (B) of FZeb. After deconvolution of the multiply charged ion signals, a mass difference of ≈35 Da was observed for the complex of dimeric CDA plus two zinc ions after incubation with FZeb, which corresponds to 0.95 water molecules per monomer of CDA. In addition, the presence of a second water molecule is revealed by the detection of species at 63,489 and 63,756 Da, corresponding to complexes consisting of dimeric CDA, two zinc ions, two water molecules, and either one or two molecules of FDHU, respectively.
When we conducted the same experiment in the presence of two equivalents of the inhibitor FZeb, the mass spectrum showed three species, ≈264 Da apart, after deconvolution (Fig. 3B). These mass increments of 264 Da are equivalent to the mass of the transition-state analogue FDHU (the covalently hydrated form of the inhibitor FZeb, expected to be formed at the active site) (Fig. 1). The relative magnitudes of these three ion signals were found to depend on the skimmer voltage applied (the potential difference between the capillary exit and the skimmer, data not shown), as expected, if the inhibitor were noncovalently bound by the enzyme. However, in experiments involving nebularization of the CDA–FZeb complex, the absolute masses from the ion signals corresponded to dimeric CDA, including two zinc atoms and two water molecules (63,226.6 Da) (Fig. 3B), and exceeded the molecular mass determined for dimeric CDA (prepared in the absence of FZeb) by 35 Da (Fig. 3A). That difference is equivalent within experimental error to the mass of two water molecules (36 Da), and can be explained by the structure of the free enzyme in Scheme 1, showing that one water molecule is bound in “split” form, with its hydroxide portion bound by Zn and its proton bound by Glu-104. The additional mass of 264 Da per active site is consistent with the view that, in the inhibitory complex, each active site contains two water molecules, one of which is covalently bound as part of FDHU, and one of which is noncovalently caught in the gap between the transition-state analogue and the surface of the active site, as shown in Scheme 2.
MS cannot exclude the alternative possibility that the protonated reaction product NH4+, with an expected mass increment of 18 Da, might be present in place of one or both of the two postulated water molecules in the enzyme–FZeb complex. However, earlier work has shown that E. coli CDA exhibits an extremely weak affinity for ammonia/ammonium ion as a competitive inhibitor (Ki > 1 M) (12), indicating that even high concentrations of ammonia/ammonium do not displace water from the active site. In the present experiments, the enzyme was nebularized from a solution containing ammonium acetate at a final concentration of 10-3 M, at least 3 orders of magnitude lower than the apparent Kd value.
Discussion
If an extra, noncovalently bound, water molecule were present in the transition-state analogue complexes that CDA forms with FDHU, and if that water molecule were sealed off from exposure to (and from equilibration with) the nearly perfect vacuum (<10-10 torr; 1 torr = 133 Pa) that is present within the chamber of the ion cyclotron, then the molecular mass of the enzyme–inhibitor complex might reveal the presence of that second water molecule. If the active site simply collapsed to fill the gap, then no excess water would be expected to be present in the inhibitor complex. In the present experiments, we sought to distinguish between these alternatives by collecting the mass spectrum of E. coli CDA in the presence and absence of a transition-state analogue inhibitor.
In the presence of FZeb, CDA remains dimeric in the vapor phase, yielding an exact mass that can be used to infer the presence of zinc, the transition-state analogue FDHU (covalently hydrated FZeb), and a single additional water molecule at each active site. The feasibility of this experiment reveals (and depends on) the surprising stability of the enzyme–inhibitor complex in the vapor phase at high vacuum. We found that the signal from this complex persisted for several seconds even at the very high vacuum (<10-10 torr) that is present within the chamber of the ion cyclotron. It appears that both the water molecule in FDHU, incorporated by the enzyme-catalyzed 3,4-addition of water to FZeb (3), and an additional water molecule that is trapped in the inhibitory complex are slow to equilibrate with the nearly perfect vacuum that surrounds the protein in the vapor phase.
Why does the dimeric enzyme nebularized in the absence of inhibitor contain no water, whereas enzyme nebularized from saturating inhibitor appear to contain a water molecule within each active site?
At equilibrium in the high vacuum present within the chamber of the spectrometer, one would expect that all water molecules, including the water molecule that is covalently bound as part of FDHU, would be lost. When the enzyme is nebularized under nondenaturing conditions in the absence of FZeb, its active site is unprotected from the nearly perfect vacuum within the chamber. We speculate that water is quickly lost from the active site, although the protein retains zinc and remains dimeric. Consistent with that possibility, the effect of solvent deuterium on kcat/Km (6) suggests that the “splitting” of water (Scheme 1) does not occur before the binding of substrate cytidine. Accordingly, it seems understandable that water appears to be lost in the absence of FZeb.
The apparent kinetic stability of the enzyme–inhibitor complex in the vapor phase, together with its water molecules, seems more surprising. That stability implies that in the absence of solvent water, the structure of the complex is maintained at least for several seconds. The apparent dissociation constant of the enzyme's complex with FZeb (10-7 M) in solution is such that this inhibitor should be released easily at low inhibitor concentrations. Moreover, dissociation would be expected to be additionally favored at high vacuum because one water molecule is produced stoichiometrically when FDHU undergoes spontaneous dehydration (Keq = 3,000) to form FZeb (5). In the presence of FZeb, we speculate that a water molecule is trapped at each active site, protected against dissociation while the inhibitory complex remains intact. The crystal structure shows that FDHU is surrounded by the active site to such an extent that <0.5% of the inhibitor is accessible to bulk solvent (4). In solution, the kinetic barrier to inhibitor release is substantial (koff ≈ 0.01 s-1; G.K.S. and M.J.S., unpublished work). If the same were true in the vapor phase, then the complex would be expected to remain largely undissociated during the few seconds that elapse between nebularization and detection in the vapor phase. Thus, it seems reasonable to suppose that the mass spectrum represents a sampling of the enzyme–inhibitor complex in the course of irreversible dissociation in the vapor phase.
Finally, the fact that the enzyme retains its affinity for FDHU in its intact, hydrated form in the vapor phase implies that the active site retains its native structure for a substantial period in the absence of solvent water. The persistence of a dimeric structure and the retention of zinc, even in the absence of FZeb, suggests that, even in the absence of the hydrophobic effect, other noncovalent forces of attraction remain in effect long enough to preserve the native structure for many seconds.
Conclusions
In this work, we tested the possibility that the presence of a water molecule might be detected by accurate MS analysis of enzymes in the presence and absence of a transition-state analogue inhibitor. In the presence of FZeb, CDA remains dimeric in the vapor phase, yielding an exact mass that can be used to infer the presence at each active site of zinc, FDHU, and a single water molecule, which remain in place for several seconds even at the very high vacuum (<10-10 torr) that is present within the chamber of the ion cyclotron. To our knowledge, this appears to be the first demonstration that MS can be used to estimate the number of water molecules in a protein, whose active site remains arguably native in the vapor phase because it continues to bind the transition-state analogue FDHU. The apparent persistence of the structure of dimeric CDA in the vapor phase indicates that the presence of solvent water is not necessary for the continued existence of enzyme structure, at least for short time intervals, in either the presence or absence of inhibitor. These findings support the view that water is retained in the inhibitory complex, as postulated earlier on the basis of entropic evidence obtained at equilibrium. A more perfect inhibitor would presumably fill that space.
Acknowledgments
This work was supported by National Institutes of Health Grants GM-18325 (to R.W.) and ES-11997 (to C.H.B.) and National Institutes of Health Training Grant GM-08570 (to G.K.S).
Author contributions: C.H.B., M.J.S., and R.W. designed research; C.H.B, G.K.S., J.P.S., and R.W. performed research; C.H.B., V.E.M., S.A.S., and R.W. contributed new reagents/analytic tools; C.H.B., M.J.S., J.P.S., and R.W. analyzed data; and C.H.B., V.E.M., G.K.S., S.A.S., M.J.S., J.P.S., and R.W. wrote the paper.
Abbreviations: CDA, cytidine deaminase; DHU, 3,4-dihydrouridine; FDHU, 5-fluoro-DHU; FZeb, 5-fluoropyrimidin-2-one ribonucleoside; FTICR, Fourier transform ion cyclotron resonance.
References
- 1.Wolfenden, R. (1972) Acc. Chem. Res. 5, 10-18. [Google Scholar]
- 2.Lienhard, G. E. (1973) Science 180, 149-155. [DOI] [PubMed] [Google Scholar]
- 3.Wolfenden, R. & Kati, W. M. (1991) Acc. Chem. Res. 23, 209-215. [Google Scholar]
- 4.Betts, L., Xiang, S., Short, S. A., Wolfenden, R. & Carter, C. W., Jr. (1994) J. Mol. Biol. 235, 635-656. [DOI] [PubMed] [Google Scholar]
- 5.Carlow, D., Short S. A. & Wolfenden, R. (1996) Biochemistry 35, 948-954. [DOI] [PubMed] [Google Scholar]
- 6.Snider, M. J., Reinhardt, L., Wolfenden, R. & Cleland, W. W. (2002) Biochemistry 41, 415-421. [DOI] [PubMed] [Google Scholar]
- 7.Snider, M. J. & Wolfenden, R. (2001) Biochemistry 40, 11364-11371. [DOI] [PubMed] [Google Scholar]
- 8.Xiang, S., Short, S. A., Wolfenden, R. & Carter, C. W., Jr. (1997) Biochemistry 36, 4768-4774. [DOI] [PubMed] [Google Scholar]
- 9.Felitsyn, N., Peschke, R. & Kebarle, P. (2002) Int. J. Mass Spectrom. 219, 39-62. [Google Scholar]
- 10.Yang, C., Carlow, D., Wolfenden, R. & Short, S. A. (1992) Biochemistry 31, 4168-4173. [DOI] [PubMed] [Google Scholar]
- 11.Smith, A. A., Carlow, D., Wolfenden, R. & Short, S. A. (1994) Biochemistry 33, 6468-6474. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, R. & Wolfenden, R. (1971) J. Biol. Chem. 246, 7561-7565. [PubMed] [Google Scholar]