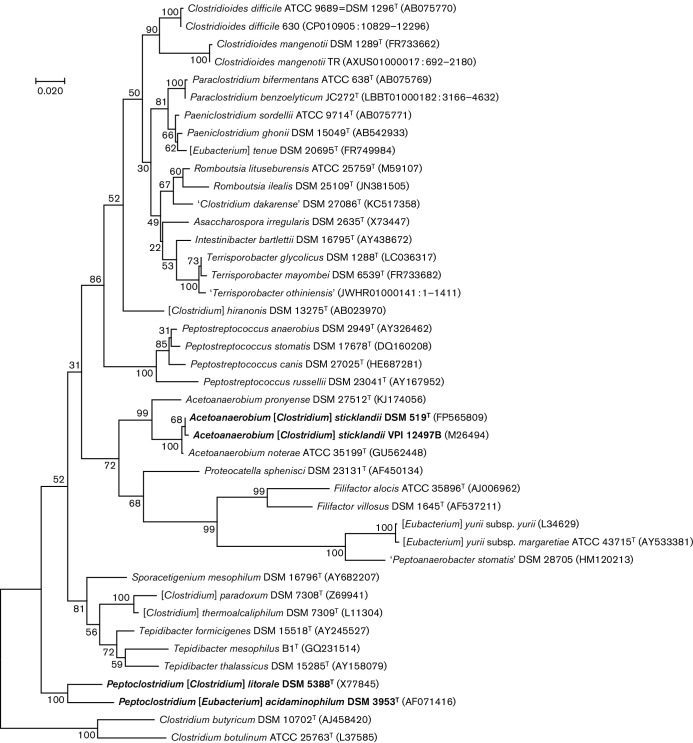
Fig. 1.
16S rRNA gene-based phylogenetic tree of the family Peptostreptococcaceae. The sequences from type strains (indicated with superscript T) were used and listed under their DSM accession numbers, where available. GenBank accession numbers are listed in parentheses. For Clostridioides difficile 630, Clostridioides mangenotii TR, Paraclostridium benzoelyticum JC272T, and ‘Terrisporobacter othiniensis’, 16S rRNA gene sequences were taken from the respective genomic entries. Quotation marks indicate the organisms whose names have not yet been validly published. The organisms that this work proposes to be renamed are indicated in boldtype. The sequences were aligned using muscle (Edgar, 2004) and the tree was inferred using the maximum-likelihood method based on the Tamura–Nei model (Tamura & Nei, 1993) as implemented in mega6 (Tamura et al., 2013); for the initial neighbour-joining tree, see Fig. S1. The tree was rooted using sequences from C. butyricum and C. botulinum, members of Clostridium sensu stricto.