Fig. 1.
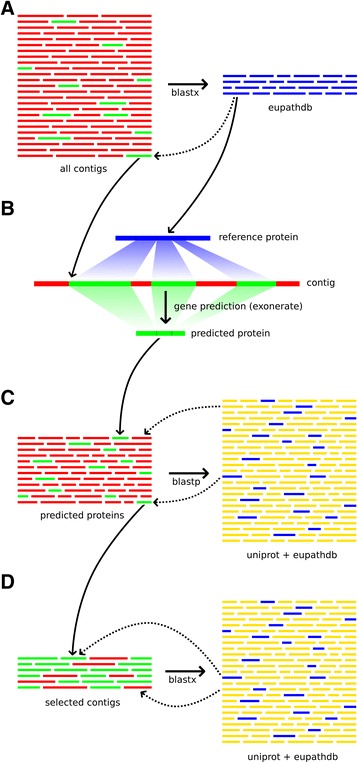
Schematic overview of the ContamFinder pipeline. a All contigs from an assembly were searched against apicomplexan proteomes from the Eukaryotic Pathogen Database (EuPathDB [19, 20]). Sequences without significant hit were discarded. b Amino acid sequences were predicted using the best hitting apicomplexan protein. Low complexity regions and repeats in the sequence were masked. c The predicted amino acid sequences were searched against the EuPathDB and UniProt database. Sequences with the best hit outside of Apicomplexa were discarded. d Unprocessed contigs corresponding to the hits from the previous step were searched against the EuPathDB and UniProt databases. Sequences that had their best hit outside of Apicomplexa were discarded. Contigs and sequence regions that were kept and used in the next step are shown in green; sequences that were discarded are denoted in red. Parasite-derived proteins in the search database are shown in blue, others in yellow
