Table 2.
WGCNA module summary and functional enrichments
| Module (gene number) | Hub genes | Viral Cor.* | Enrichment terms | Immune cell type enrichment |
|---|---|---|---|---|
Pink (100) 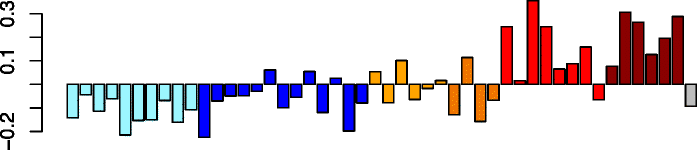
|
SPI1 | 0.83 | GO:0006952 defense response (P adj = 6.17e-07) | Neutrophils (P = 7.46e-11) |
| PLEK | GO:0006955 immune response (P adj = 2.25e-06) | Act. neutrophils (P = 4.44e-09) | ||
| HCK | IPA path: Fcγ receptor-mediated phagocytosis in macrophages and monocytes (P = 4.97e-10) | Eosinophils (P = 3.74e-04) | ||
| FGR | IPA upstream regulator: IFNG (P = 4.77e-16) | Act. macrophages (P = 9.78e-03) | ||
| TLR4 | IPA function: leukocyte migration (increased) (P = 3.82e-22) | Act. eosinophils (P = 2.67e-02) | ||
Blue (726) 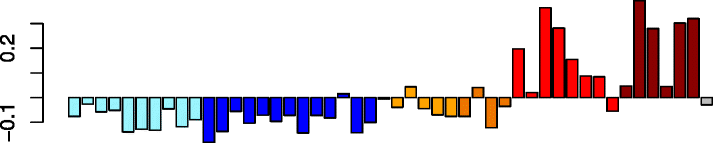
|
SLAMF7 | 0.75 | GO:0006955 immune response (P adj = 5.31e-76) | Act. macrophages (P = 2.49e-16) |
| SP100 | GO:0006952 defense response (P adj = 2.15e-29) | Dendritic cells (P = 3.01e-11) | ||
| DTX3L | GO:0045321 leukocyte activation (P adj = 3.79e-26) | Act. neutrophils (P = 3.78e-05) | ||
| GIMAP4 | IPA upstream regulator: IFNG (P = 1.66e-100) | Neutrophils (P = 1.99e-04) | ||
| OAS3 | IPA path: interferon signaling (P = 3.35e-22) | NK cells (P = 2.64e-02) | ||
Black (117) 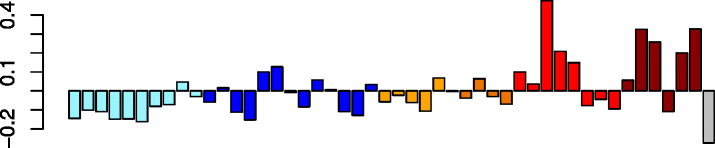
|
DCAKD | 0.28 | GO:0031982 ~ vesicle (P adj = 1.38e-02) | Act. macrophages (P = 4.06e-03) |
| GNA13 | GO:0012505 endomembrane system (P adj = 2.10e-02) | |||
| PICALM | IPA upstream regulator: CHEK2 (P = 6.49e-05) | |||
| CREB1 | IPA path: ephrin receptor signaling (P = 3.26e-04) | |||
| LARP4 | IPA function: viral infection (increased) (P = 1.02e-04) | |||
Magenta (85) 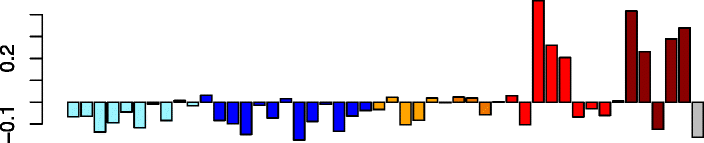
|
TNKS2 | 0.32 | No significant DAVID enrichments | Act. macrophages (P = 4.57e-02) |
| VCPIP1 | IPA path: oxidative phosphorylation (P = 8.05e-05) | |||
| MIER1 | IPA path: mitochondrial dysfunction (P = 6.51e-04) | |||
| CD2AP | IPA path: NF-κB signaling (P = 6.14e-03) | |||
| ARAP2 | ||||
Yellow (189) 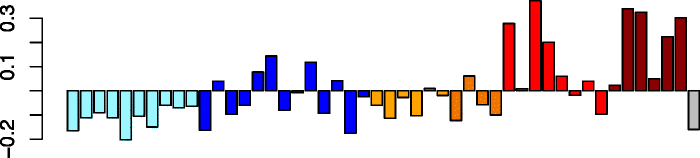
|
CHST9 | 0.70 | No significant DAVID enrichments | Act. neutrophils (P = 4.00e-04) |
| ITGA5 | IPA function: inflammatory response (P = 2.09e-11) | Act. macrophages (P = 9.78e-03) | ||
| C8orf47 | IPA function: quantity of leukocytes (P = 1.67e-10) | Neutrophils (P = 1.56e-02) | ||
| OXSR1 | IPA function: differentiation of cells (P = 1.74e-10) | Act. eosinophils (P = 1.83e-02) | ||
| ITPRIPL2 | ||||
Green (179) 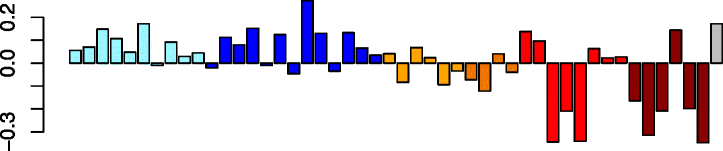
|
RPL10A | −0.46 | GO:0006414 translational elongation (P adj = 3.65e-44) | B cells (P adj = 1.07e-07) |
| RPL3 | KEGG: hsa03010 ribosome (P adj = 3.34e-37) | Th2 cells (P adj = 8.30e-05) | ||
| EEF2 | GO:0006412 translation (P adj = 7.72e-36) | T cells (P adj = 2.33e-04) | ||
| RPS14 | IPA path: eIF2 signaling (P = 4.07e-47) | Mast cells (P adj = 1.33e-03) | ||
| RPL4 | Th1 cells (P adj = 2.42e-02) | |||
Brown (203) 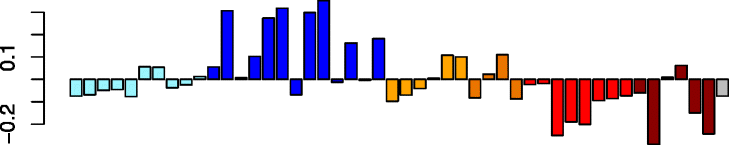
|
ELOVL5 | −0.74 | Type 2 inflammation (P = 3.30e-04) | No significant immune cell enrichments |
| CDH26 | No significant DAVID enrichments | |||
| ALOX15 | IPA upstream regulator: IL13 (P = 2.77e-05) | |||
| FETUB | ||||
| VWF | ||||
Red (164) 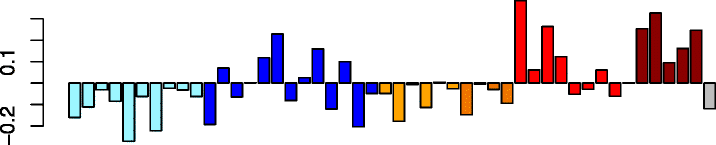
|
CFL1 | 0.50 | GO:0015629 actin cytoskeleton (P adj = 2.41e-06) | Act. neutrophils (P adj = 2.35e-02) |
| CAP1 | GO:0031252 cell leading edge (P adj = 6.68e-05) | Act. macrophages (P adj = 3.04e-02) | ||
| VASP | IPA path: remodeling of epithelial adherens junctions (P = 2.42e-09) | |||
| DIAPH1 | IPA upstream predictor: TGFB1 (P = 6.88e-09) | |||
| TMBIM1 | IPA function: organization of cytoskeleton (increased) (P = 3.65e-15) | |||
Turquoise (1,615) 
|
C6orf165 | −0.43 | GO:0005929 cilium (P adj = 2.78e-30) | No significant immune cell enrichments |
| ARMC2 | GO:0015630 microtubule cytoskeleton (P adj = 1.02e-23) | |||
| CAPSL | GO:0005930 axoneme (P = 3.01e-21) | |||
| EFCAB6 | IPA upstream regulator: RFX3 (P = 1.74e-05) | |||
| ANKRD66 | IPA function: formation of cilia (decreased) (P = 1.47e-43) |
*Cor = Spearman correlation of module eigengenes with depth of detected viral genomes (for virus high samples only)
