Abstract
The microRNA, miR-196b, serves a role in normal cell differentiation, proliferation and tumorigenesis of different types of cancer. The aim of the present study was to explore the serum expression of miR-196b in colorectal cancer (CRC) and its correlation with clinicopathological features. Sera samples were obtained from 103 patients with CRC, 51 patients with colorectal adenoma (Ad) and 100 healthy individuals for the present study. The serum expression of miR-196b in sera samples of the three cohorts was detected using reverse transcription-quantitative polymerase chain reaction. The diagnostic value of miR-196b in the serum of the patients with CRC was evaluated by receiver operating characteristic (ROC) curve and survival analysis, using the Kaplan-Meier method, which was performed with the data from a 5-year follow-up. The expression of miR-196b in the serum of patients with CRC was significantly higher compared with that in Ad patients or healthy individuals (all P<0.001), and the overexpression of serum miR-196b was clearly associated with lymph node invasion, differentiation, and the tumor-lymph nodes-metastasis stage (all P<0.05). ROC curve analysis demonstrated that, comparing patients with CRC with healthy individuals, the area under the curve of serum miR-196b was 0.8135, and its specificity and sensitivity were 63 and 87.38%, respectively, at a diagnostic threshold of −4.785. Patients with CRC of miR-196b-high status had shorter overall survival and disease-free survival rates compared with those of miR-196b-low status. In conclusion, the results of the present study demonstrated that serum miR-196b is upregulated in CRC, and may have an application as a diagnostic and prognostic biomarker for patients with CRC.
Keywords: miR-196b, biomarker, colorectal cancer, diagnosis, prognosis
Introduction
Colorectal cancer (CRC), one of most prevalent malignancies, is considered to be the third most commonly diagnosed cancer (1.36 million cases), and the fourth cause of cancer-associated mortality (694,000 deaths) worldwide (1). Colorectal adenoma (Ad), also known as one of the pre-cancerous lesion types, is closely associated with CRC, and is present in the majority of cases of CRC (2). In spite of what has thus far been accomplished in terms of diagnosing and treating CRC, the diagnostic rate of CRC requires further improvement, and the prognosis of patients with advanced CRC remains poor. Therefore, it is crucial to explore the underlying mechanisms of carcinogenesis in CRC and to identify novel, sensitive and specific diagnostic biomarkers to improve diagnostic efficiency in cases of CRC.
MicroRNAs (miRNAs or miRs) are a class of small, evolutionarily conserved, non-coding single-stranded nucleotide molecules (20–24 nt), which are able to inhibit or promote gene expression via binding to the 3′-untranslated region of the target messenger RNA (mRNA) at the post-transcriptional level (3–5). A burgeoning body of evidence has demonstrated that miRNAs serve a crucial role in a variety of biological processes, including cell growth (6), cell apoptosis (7) and development of the nervous system (8). In oncological studies, a growing number of miRNAs have been confirmed to have an association with a large number of neoplasms (9–11). For example, miR-15a and miR-16 may be closely associated with cancer pathogenesis (12,13). miR-143, as a tumor suppressor, inhibits the growth, and induces apoptosis of, gastric cancer cells through targeting cyclooxygenase-2 (14). To date, miRNAs have been detected and extracted not only in tissues, but also in the plasma, serum and urine (15). Willeit et al (16) demonstrated that miR-122, a liver miRNA, is able to function as a novel biomarker for cardiovascular and metabolic diseases. Therefore, miRNAs, as regulators of gene expression, are involved in the pathogenesis of numerous types of tumors, and are able to be applied as potential biomarkers for tumor diagnosis.
The miR-196 family, which is most strongly correlated with malignancies, contains three members, miR-196a-1, miR-196a-2 and miR-196b, which have been reported to be involved in several biological processes, including embryonic development and neoplasia (3,17,18). Emerging evidence has indicated that the aberrant expression of miR-196b is closely associated with leukemogenesis via increasing the population of leukemic stem/progenitor cells, blocking cell differentiation, promoting cell proliferation and diminishing cell apoptosis (19). Furthermore, Ge et al (20) demonstrated that expression of miR-196b in tissues may have a significant correlation with an aggressive progression of the disease and poor clinical outcomes in patients with CRC. However, the association between serum expression of miR-196b and CRC has yet to be fully elucidated.
The present study, has sought to determine whether the expression of serum miR-196b is upregulated in CRC, and to evaluate the diagnostic value of serum miR-196b in CRC.
Materials and methods
Ethics statement
The present study was approved by the Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University (Shanghai, China). All examinations were performed after obtaining written informed consent from patients and volunteers.
Patients and serum
Between February 2010 and February 2015, 103 patients with primary CRC, comprising 62 males and 41 females, who accepted surgical resection in Renji Hospital, School of Medicine, Shanghai Jiao Tong University, were recruited in the present study. All the patients were diagnosed with CRC by two experienced pathologists, and did not receive chemotherapy or radiotherapy. In addition, 51 patients with Ad and 100 healthy individuals were enrolled as a middle group and the control group, respectively. Baseline information of all the groups is shown in Table I. Blood samples (5 ml) from the 103 patients with CRC, 51 patients with Ad and 100 healthy controls were collected in tubes containing ethylenediaminetetra-acetic acid (EDTA) prior to surgical operation. The sera samples were separated by centrifugation in two successive steps: A centrifugation at 1,600 × g for 10 min at 4°C, followed by a second centrifugation at 16,000 × g for 10 min at 4°C. Subsequently, the supernatant sera were stored in liquid nitrogen at −80°C for reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Table I.
Patient characteristics in the three groups.
| Characteristic | CRC group, n=103 | Ad group, n=51 | Control group, n=100 | P-value | |
|---|---|---|---|---|---|
| Age, years | 52.8±4.6 | 54.3±5.1 | 53.4±4.8 | 0.562 | |
| Gender | |||||
| Male | 62 (60.2%) | 29 (56.9%) | 52 (52.0%) | 0.712 | |
| Female | 41 (39.8%) | 22 (43.1%) | 48 (48.0%) | ||
| Nation | |||||
| Han Chinese | 95 (92.2%) | 49 (96.1%) | 94 (94.0%) | 0.664 | |
| Minority | 8 (7.2%) | 2 (3.9%) | 6 (6.0%) | ||
| Registered residence | |||||
| Urban | 64 (62.1%) | 26 (51.0%) | 56 (56.0%) | 0.390 | |
| Rural | 39 (37.9%) | 25 (49.0%) | 44 (44.0%) | ||
| History of alcohol consumption | |||||
| Yes | 60 (58.3%) | 23 (45.1%) | 52 (52.0%) | 0.293 | |
| No | 43 (41.7%) | 28 (54.9%) | 48 (48.0%) | ||
| History of smoking | |||||
| Yes | 47 (45.6%) | 27 (52.9%) | 48 (48.0%) | 0.694 | |
| No | 56 (54.4%) | 24 (47.1%) | 52 (52.0%) |
CRC, colorectal cancer; Ad, adenoma.
RNA isolation and RT-qPCR
Total RNA was extracted from 100 µl serum using a Qiagen miRNeasy Mini kit (Qiagen, Valencia, CA, USA). The cDNA of miR-196b was reverse-transcribed from total RNA using a PrimeScript RT-PCR kit (Takara Biotechnology Co., Ltd., Dalian, China). U6 RNA was used as an internal control in the present study. The four primers used in this study are listed in Table II. PCR for detecting the expression levels of miR-196b was performed using an Applied Biosystems 7900 QPCR system (Applied Biosystems, Foster City, CA, USA) with a 20 µl RT-qPCR reaction mixture, comprising forward primer (0.6 µl), reverse primer (0.6 µl), cDNA (2 µl), ROX Reference Dye II (0.4 µl), SYBR Premix Ex Taq (10 µl), and doubly distilled H2O (6.6 µl). The relative expression of miRNA-196b was calculated using the 2−ΔΔCq method (21), with U6 RNA as the internal reference compound.
Table II.
Primers for performing reverse transcription-quantitative polymerase chain reaction on miR-196b and U6.
| Gene | Primer sequence |
|---|---|
| miR-196b | |
| Forward | 5′-TAGGTACCACTTTATCCCGTTCACCA-3′ |
| Reverse | 5′-ATCTCGAGGCAGGGAGAGAGGAATAA-3′ |
| U6 | |
| Forward | 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse | 5′-AACGCTTCACGAATTTGCGT-3′ |
Receiver operating characteristic (ROC) curve analysis
The relative levels of miR-196 expression in the serum of patients and controls were recorded. ROC curves were generated, with the horizontal axis as specificity and the vertical axis as sensitivity, using GraphPad Prism 5 software (GraphPad Software, San Diego, CA, USA). According to the drawn ROC curves, the diagnosis cut-off points and their specificity and sensitivity were analyzed and calculated. The diagnostic value of miR-196b in the serum in CRC was evaluated, and presented by the area under the curve (AUC) and the standard error.
Survival analysis
Patients in the CRC group were divided into two subgroups on the basis of whether the expression of miR-196b was high or low. Survival analysis using the Kaplan-Meier method was performed, using the data from a 5-year follow-up.
Statistical analysis
All data were performed using SPSS 19.0 software (IBM SPSS, Armonk, NY, USA). Values are expressed as the mean ± standard deviation. Differences and/or correlations between groups were analyzed and calculated using the Student's t-test and Chi-square test. Survival curves were drawn using the Kaplan-Meier method, and the long-rank test was employed for survival analysis. Prognosis analysis was performed using multivariate cox proportional hazards regression analysis, and P<0.05 was considered to indicate a statistically significant value.
Results
Serum miR-196b is upregulated in the CRC group compared with the Ad group and healthy individuals, and is a potential diagnostic marker for CRC
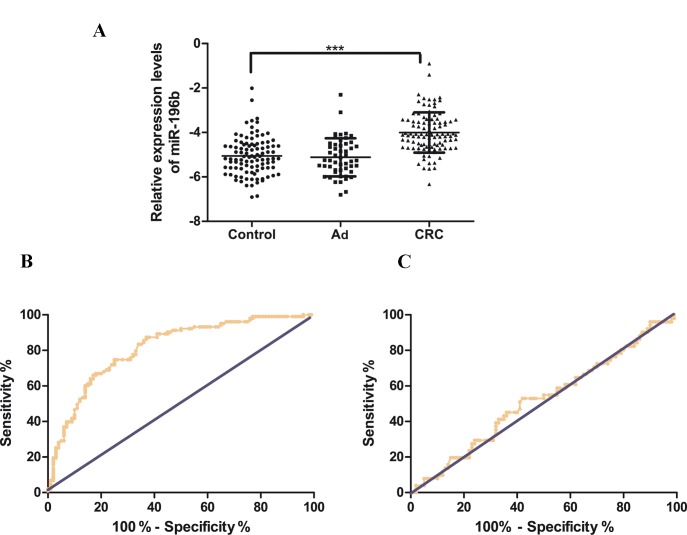
Serum miR-196b was detected in the three groups of participants, including healthy individuals (n=100), Ad (n=51) and CRC (n=103). As shown in Fig. 1A, compared with healthy controls, the serum level of miR-196b was significantly upregulated in CRC (CRC vs. controls: −3.91±0.89 vs. −5.10±0.85, P<0.001). However, the serum miR-196b levels were slightly downregulated in Ad compared with the healthy controls, although without statistical significance (Ad vs. controls: −5.12±0.86 vs. −5.10±0.85, P=0.68). These results indicated that the serum expression of miR-196b was significantly upregulated in CRC. ROC curves were drawn to analyze and assess the diagnostic power of serum miR-196b in CRC (Fig. 1B). The AUC was 0.8135 (95% confidence interval: 0.7546–0.8725), with a diagnostic threshold of −4.785, and specificity and sensitivity were 87.38 and 63%, respectively. However, as shown in Fig. 1C, serum miR-196b had no diagnostic value in the Ad group.
Figure 1.
Serum miR-196b expression is decreased in CRC. (A) Levels of serum miR-196b are shown. (B) The respective area under receiver operating characteristic curves for CRC. (C) The respective area under receiver operating characteristic curves for Ad. Ad, adenoma; CRC, colorectal cancer.
Association between the clinicopathological features and expression levels of serum miR-196b in human CRC
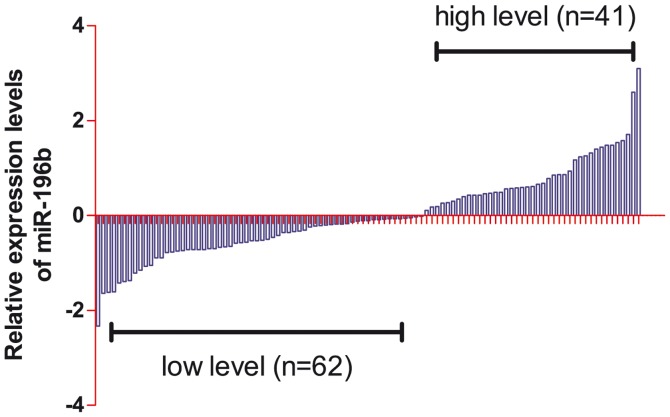
As shown in Fig. 2, relative miR-196b levels detected by RT-qPCR in the serum of the 103 patients with CRC were divided into the two subgroups on the basis of the expression levels of miR-196b: The high serum expression of miR-196b group, and the low serum expression of miR-196b group. The correlations between clinicopathological features and serum miR-196b levels are shown in Table III. The results of the chi-square test demonstrated that low expression of plasma miR-145 was significantly associated with lymph node invasion, differentiation and tumor-lymph nodes-metastasis (TNM) staging (all P<0.05), whereas no significant associations were determined for age, gender, history of alcohol consumption, tumor size, tumor invasion, metastasis and site of the primary tumor (all P>0.05).
Figure 2.
Relative serum miR-196b expression in the colorectal cancer group (n=103). For each column, a value <0 is defined as low expression, and values >0 are defined as high expression.
Table III.
Associations between serum miR-196b levels and clinicopathological parameters in patients with CRC.
| Serum miR-196b expression | ||||
|---|---|---|---|---|
| Characteristic | Total no. | Low (n=62) | High (n=41) | P-value |
| Age | ||||
| <57 | 45 | 30 (44.6%) | 15 (41.4%) | 0.237 |
| ≥57 | 58 | 32 (55.4%) | 26 (48.6%) | |
| Gender | ||||
| Male | 62 | 34 (57.1%) | 28 (63.8%) | 0.172 |
| Female | 41 | 28 (42.9%) | 13 (36.2%) | |
| History of alcohol consumption | ||||
| Yes | 47 | 29 (67.9%) | 18 (60.3%) | 0.775 |
| No | 56 | 33 (32.1%) | 23 (39.7%) | |
| Tumor size | ||||
| <5 cm | 59 | 38 (26.8%) | 21 (48.3%) | 0.254 |
| ≥5 cm | 44 | 24 (73.2%) | 20 (51.7%) | |
| Tumor invasion | ||||
| T1-T2 | 51 | 27 (52.9%) | 23 (47.1%) | 0.212 |
| T3-T4 | 51 | 35 (68.6%) | 18 (31.4%) | |
| Metastasis | ||||
| Yes | 33 | 22 (60.7%) | 11 (41.4%) | 0.357 |
| No | 70 | 40 (39.3%) | 30 (58.6%) | |
| Lymph node invasion | ||||
| Yes | 45 | 34 (67.9%) | 11 (56.9%) | 0.005 |
| No | 58 | 28 (32.1%) | 30 (43.1%) | |
| Differentiation | ||||
| Well | 21 | 15 (12.5%) | 6 (%) | 0.001 |
| Moderate | 43 | 32 (26.8%) | 11 (43.1%) | |
| Poor | 39 | 15 (60.7%) | 24 (44.8%) | |
| TNM stage | ||||
| I–II | 57 | 42 (19.0%) | 15 (41.4%) | 0.002 |
| III–IV | 46 | 20 (81.0%) | 26 (58.6%) | |
| Site of primary tumor | ||||
| Right-sided | 40 | 25 | 15 | 0.875 |
| Left-sided | 42 | 24 | 18 | |
| Rectum | 21 | 13 | 8 | |
TNM, tumor-lymph nodes-metastasis. The P-values shown in bold are the statistically significant values (P<0.05).
Prognostic effect of upregulation of serum miR-196b in human CRC
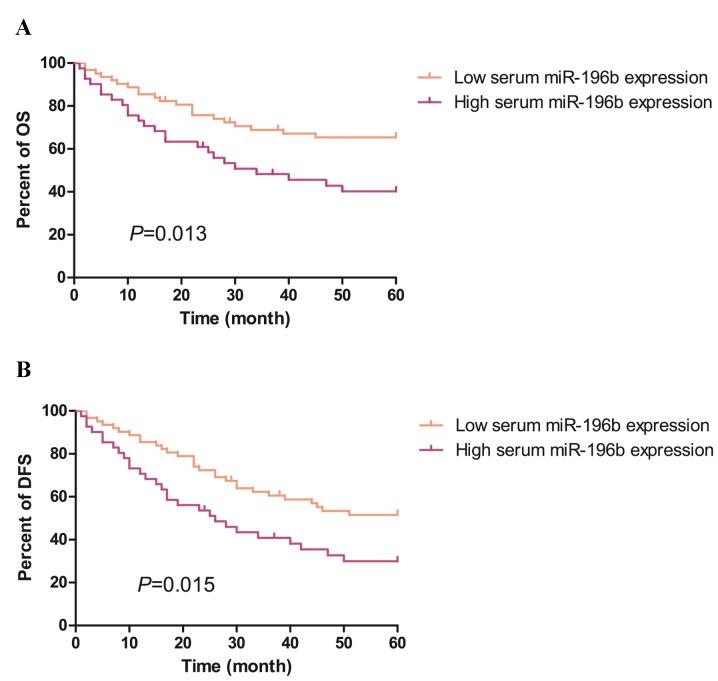
Subsequently, the prognostic implications of an increased expression of serum miR-196b in CRC was investigated using the Kaplan-Meier method. As shown in Fig. 3A and B, survival analysis indicated that overall survival (OS) and disease-free survival (DFS) rates of patients with CRC with high serum miR-196b levels were significantly lower than those of patients with low serum miR-196b levels (OS: 40.2 vs. 65.4, P<0.05; DFS: 40.2 vs. 65.4, P<0.05). Therefore, serum miR-196b was a prognostic indicator for DFS and OS of patients with CRC.
Figure 3.
Prognostic value of serum miR-196b in colorectal cancer. (A) Kaplan-Meier survival curves of the DFS. (B) Kaplan-Meier survival curves of OS. P<0.05 was considered to indicate statistically significant values. DFS, disease-free survival; OS, overall survival.
Multivariate Cox's proportional hazard regression analysis of the prognostic values of indicators in human CRC
It remains unclear whether the independent parameters of serum miR-196b expression in the prognosis of human CRC are correlated with OS. In the present study, in order to explore the associations between the independent predictors of serum miR-196b expression in the prognosis of human CRC and OS, the clinical characteristics, including serum miR-196b expression, lymph node invasion, differentiation and TNM stage, which were defined as statistically significant, were performed using multivariate Cox's proportional hazard regression analysis. According to the multivariate analysis, the serum miR-196b expression, lymph node invasion and TNM stage were all identified as having significant associations with OS. Therefore, the present investigation has shown that independent predictors, including serum miR-196b expression, lymph node invasion and TNM stage, are potential risk factors for OS in human CRC (all P<0.05; Table IV).
Table IV.
Multivariate Cox's proportional hazards regression analysis of the prognostic values of indicators on overall survival in human CRC.
| 95% CI for Exp (B) | |||||
|---|---|---|---|---|---|
| Characteristic | Unfavorable vs. favorable | P-value | Exp (B) | Lower | Upper |
| Serum miR-196b level | High vs. low | 0.045 | 2.8 | 1.023 | 7.664 |
| Lymph node invasion | Yes vs. no | 0.013 | 0.256 | 0.088 | 0.749 |
| Differentiation status | Poor vs. well, moderate | 0.431 | 1.468 | 0.565 | 3.814 |
| TNM stage | III–IV vs. I–II | 0.022 | 3.31 | 1.193 | 9.185 |
B, regression coefficient; Exp (B), index of the regression coefficients; CI, confidence interval; TNM, tumor-lymph nodes-metastasis. The P-values shown in bold are the statistically significant values (P<0.05).
Discussion
At the present time, the mortality rate due to CRC remains higher than that for a number of other common malignancies. Recently, a burgeoning list of tumor biomarkers, including MYC, HOTAIRM1 and colon cancer-associated transcript 1 (CCAT1), have been reported to have a limited association with diagnosis and prognosis in CRC (22–24). Therefore, it is of great interest to identify further, more specific and sensitive biomarkers for the diagnosis of CRC. In the present study, it was noted that, compared with healthy individuals, the expression of serum miR-196b was significantly upregulated in patients with CRC, and serum miR-196b was identified to be a potential diagnostic marker for CRC. Furthermore, Cox regression analysis revealed that serum miR-196b expression, lymph node invasion, differentiation and the TNM stage were potential risk factors for OS in human CRC.
Tissue miRNAs, as the biomarkers of diagnosis, are most widely used in scientific research. Fadous-Khalifé et al (25) demonstrated that, compared with normal tissue, the expression of Kruppel-like factor 4 (KLF4) was markedly decreased in non-small cell lung cancer, although it was overexpressed in small cell lung cancer. In addition, Perini et al (26) demonstrated that the overexpression of membrane epidermal growth factor receptor is associated with decreased survival in pancreas cancer tissue. However, patients with a number of different types of cancer are unable to undergo surgery when they are diagnosed. The diagnostic effectiveness of this method is unsatisfactory, due to the limitation of there being a dependence on surgical resection, and the invasive procedure for the collection of tissue samples.
Given the great advances that have been made in diagnostic technology, with the exception of that associated with the tissues, it is possible that the expression of circulating miRNAs, which can be detected in, and extracted from, a variety of biological samples, including serum, plasma and urine, could be a useful addition to the technologies available. The miR-196 family of miRNAs, which is located at three paralogous loci in the mammalian homeobox (HOX) clusters, is able to regulate a series of genetic processes in the development of embryos (27). As a member of the miR-196 family, the aberrant expression of miR-196b was determined to be associated with a great number of cancer types, including lung, oral and gastric cancers, and acute myeloid leukemia (28–31). Low expression of miR-196b leads to the development of chronic myeloid leukemia via upregulation of the expression of the BCR-ABL1 fusion gene and HOXA9 (32). In addition, How et al (33) revealed that the low expression of miR-196b was significantly correlated with poorer DFS rates for patients with cervical cancer, and that the recovery of miR-196b expression resulted in reduced tumor angiogenesis and tumor cell proliferation in vivo, and reduced cell growth, clonogenicity, migration and invasion in vitro. Furthermore, Li et al (34) demonstrated that miR-196b expression was clearly associated with mixed lineage leukemia-rearranged leukemia by directly targeting the HOXA9/MEIS1 oncogenes and the FAS tumor suppressor. In contrast, upregulation of miR-196b expression was identified in glioblastoma, which was associated with a poor prognosis via promoting cellular proliferation (35). A similar result in gastric cancer revealed that, via regulation of the phosphoinositide 3-kinase/AKT/mammalian target of rapamycin pathway, overexpression of miR-196b promotes the proliferation and invasion of gastric cancer cells (36). Additionally, Wang et al (37) demonstrated that miR-196b was upregulated in colonic cancer tissues. Although the phenomenon of the aberrant expression of miR-196b in CRC is partly understood, little is known about the association between serum miR-196b expression and the clinical relevance of such an aberration. In the present study, based on three large cohorts, including CRC, Ad and healthy individuals, we have demonstrated that serum miR-196b was significantly upregulated in the CRC group compared with the Ad and the healthy individuals groups. It was also demonstrated that high levels of miR-196b in CRC sera were associated with positive lymph node invasion, the TNM stage and poor differentiation. Furthermore, miR-196b-high status was clearly correlated with shorter OS and DFS rates compared with those with miR-196b-low status. Finally, the Cox regression analysis indicated that high serum miR-196b expression, lymph node invasion and the TNM stage were risk factors for OS in human CRC.
The present study did have several limitations. First, this study could be repeated with the analysis and evaluation of a larger number of patients, which may help to improve the accuracy of the data and credibility of the study. Secondly, there may have been selection bias in operation when the inclusion criteria were formulated.
In conclusion, the present study has revealed that the serum expression of miR-196b was significantly higher in patients with CRC compared with that in Ad patients or healthy individuals. In addition, the results of our study have indicated that the overexpression of serum miR-196b was significantly correlated with lymph node invasion, the TNM stage, poor differentiation and poor prognosis. Therefore, serum miR-196b may have an application as a diagnostic tool and prognostic marker for CRC.
Acknowledgements
Τhe present study was supported by the following support fund: The Shanghai Municipal Commission of Health and Family Planning Fund (grant no. 20154Y0207).
References
- 1.Wei Q, Wang X, Gao J, Li J, Li J, Qi C, Li Y, Li Z, Shen L. Clinicopathologic and molecular features of colorectal adenocarcinoma with signet-ring cell component. PLoS One. 2016;11:e0156659. doi: 10.1371/journal.pone.0156659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strum WB. Colorectal Adenomas. N Engl J Med. 2016;374:1065–1075. doi: 10.1056/NEJMra1513581. [DOI] [PubMed] [Google Scholar]
- 3.Fantini S, Salsi V, Vitobello A, Rijli FM, Zappavigna V. MicroRNA-196b is transcribed from an autonomous promoter and is directly regulated by Cdx2 and by posterior Hox proteins during embryogenesis. Biochim Biophys Acta. 2015;1849:1066–1080. doi: 10.1016/j.bbagrm.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Kojima S, Goto Y, Naya Y. The roles of microRNAs in the progression of castration-resistant prostate cancer. J Hum Genet. 2016 Jun 9; doi: 10.1038/jhg.2016.69. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 5.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: Playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Zheng K, Li H, Huang H, Qiu M. MicroRNAs and glial cell development. Neuroscientist. 2012;18:114–118. doi: 10.1177/1073858411398322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lizé M, Klimke A, Dobbelstein M. MicroRNA-449 in cell fate determination. Cell Cycle. 2011;10:2874–2882. doi: 10.4161/cc.10.17.17181. [DOI] [PubMed] [Google Scholar]
- 8.Cao DD, Li L, Chan WY. MicroRNAs: Key Regulators in the Central Nervous System and Their Implication in Neurological Diseases. Int J Mol Sci. 2016;17:E842. doi: 10.3390/ijms17060842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rea D, Del Vecchio V, Palma G, Barbieri A, Falco M, Luciano A, De Biase D, Perdonà S, Facchini G, Arra C. Mouse Models in Prostate Cancer Translational Research: From Xenograft to PDX. Biomed Res Int. 2016;2016:9750795. doi: 10.1155/2016/9750795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landi D, Moreno V, Guino E, Vodicka P, Pardini B, Naccarati A, Canzian F, Barale R, Gemignani F, Landi S. Polymorphisms affecting micro-RNA regulation and associated with the risk of dietary-related cancers: A review from the literature and new evidence for a functional role of rs17281995 (CD86) and rs1051690 (INSR), previously associated with colorectal cancer. Mutat Res. 2011;717:109–115. doi: 10.1016/j.mrfmmm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Skrzypski M, Dziadziuszko R, Jassem J. MicroRNA in lung cancer diagnostics and treatment. Mutat Res. 2011;717:25–31. doi: 10.1016/j.mrfmmm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Acunzo M, Croce CM. Downregulation of miR-15a and miR-16-1 at 13q14 in Chronic Lymphocytic Leukemia. Clin Chem. 2016;62:655–656. doi: 10.1373/clinchem.2015.240036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang E, Liu R, Chu Y. miRNA-15a/16: as tumor suppressors and more. Future Oncol. 2015;11:2351–2363. doi: 10.2217/fon.15.101. [DOI] [PubMed] [Google Scholar]
- 14.Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q, Dan ZL, Tian DA, Zhang P. MicroRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J Gastroenterol. 2013;19:7758–7765. doi: 10.3748/wjg.v19.i43.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marí-Alexandre J, Sánchez-Izquierdo D, Gilabert-Estellés J, Barceló-Molina M, Braza-Boïls A, Sandoval J. miRNAs Regulation and Its Role as Biomarkers in Endometriosis. Int J Mol Sci. 2016;17:E93. doi: 10.3390/ijms17010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willeit P, Skroblin P, Kiechl S, Fernandez-Hernando C, Mayr M. Liver microRNAs: potential mediators and biomarkers for metabolic and cardiovascular disease? Eur Heart J. 2016 Apr 20; doi: 10.1093/eurheartj/ehw146. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 18.Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16:4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 19.Pan Y, Meng M, Zhang G, Han H, Zhou Q. Oncogenic microRNAs in the genesis of leukemia and lymphoma. Curr Pharm Des. 2014;20:5260–5267. doi: 10.2174/1381612820666140128211724. [DOI] [PubMed] [Google Scholar]
- 20.Ge J, Chen Z, Li R, Lu T, Xiao G. Upregulation of microRNA-196a and microRNA-196b cooperatively correlate with aggressive progression and unfavorable prognosis in patients with colorectal cancer. Cancer Cell Int. 2014;14:128. doi: 10.1186/s12935-014-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Lee KS, Kwak Y, Nam KH, Kim DW, Kang SB, Choe G, Kim WH, Lee HS. c-MYC Copy-Number Gain Is an Independent Prognostic Factor in Patients with Colorectal Cancer. PLoS One. 2015;10:e0139727. doi: 10.1371/journal.pone.0139727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M, Zhang H. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016;20:2036–2044. doi: 10.1111/jcmm.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xin Y, Li Z, Shen J, Chan MT, Wu WK. CCAT1: A pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016;49:255–260. doi: 10.1111/cpr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fadous-Khalifé MC, Aloulou N, Jalbout M, Hadchity J, Aftimos G, Paris F, Hadchity E. Krüppel-like factor 4: A new potential biomarker of lung cancer. Mol Clin Oncol. 2016;5:35–40. doi: 10.3892/mco.2016.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perini MV, Montagnini AL, Coudry R, Patzina R, Penteado S, Abdo EE, Diniz A, Jukemura J, da Cunha JE. Prognostic significance of epidermal growth factor receptor overexpression in pancreas cancer and nodal metastasis. ANZ J Surg. 2015;85:174–178. doi: 10.1111/ans.12399. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Lindsay J, Feng ZP, Frankenberg S, Hu Y, Carone D, Shaw G, Pask AJ, O'Neill R, Papenfuss AT, et al. Evolution of coding and non-coding genes in HOX clusters of a marsupial. BMC Genomics. 2012;13:251. doi: 10.1186/1471-2164-13-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tellez CS, Juri DE, Do K, Picchi MA, Wang T, Liu G, Spira A, Belinsky SA. miR-196b Is Epigenetically Silenced during the Premalignant Stage of Lung Carcinogenesis. Cancer Res. 2016;76:4741–4751. doi: 10.1158/0008-5472.CAN-15-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou YY, You JJ, Yang CM, Pan HW, Chen HC, Lee JH, Lin YS, Liou HH, Liu PF, Chi CC, et al. Aberrant DNA hypomethylation of miR-196b contributes to migration and invasion of oral cancer. Oncol Lett. 2016;11:4013–4021. doi: 10.3892/ol.2016.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li CY, Liang GY, Yao WZ, Sui J, Shen X, Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al. Identification and functional characterization of microRNAs reveal a potential role in gastric cancer progression. Clin Transl Oncol. 2016 May 12; doi: 10.1007/s12094-016-1516-y. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 31.Díaz-Beyá M, Brunet S, Nomdedéu J, Tejero R, Díaz T, Pratcorona M, Tormo M, Ribera JM, Escoda L, Duarte R, et al. Cooperative AML group CETLAM (Grupo Cooperativo Para el Estudio y Tratamiento de las Leucemias Agudas y Mielodisplasias): MicroRNA expression at diagnosis adds relevant prognostic information to molecular categorization in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leukemia. 2014;28:804–812. doi: 10.1038/leu.2013.281. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Zheng W, Song Y, Ma W, Yin H. Low expression of miR-196b enhances the expression of BCR-ABL1 and HOXA9 oncogenes in chronic myeloid leukemogenesis. PLoS One. 2013;8:e68442. doi: 10.1371/journal.pone.0068442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.How C, Hui AB, Alajez NM, Shi W, Boutros PC, Clarke BA, Yan R, Pintilie M, Fyles A, Hedley DW, et al. MicroRNA-196b regulates the homeobox B7-vascular endothelial growth factor axis in cervical cancer. PLoS One. 2013;8:e67846. doi: 10.1371/journal.pone.0067846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma R, Yan W, Zhang G, Lv H, Liu Z, Fang F, Zhang W, Zhang J, Tao T, You Y, et al. Upregulation of miR-196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS One. 2012;7:e38096. doi: 10.1371/journal.pone.0038096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li NA, Wang W, Xu B, Gong H. miR-196b regulates gastric cancer cell proliferation and invasion via PI3K/AKT/mTOR signaling pathway. Oncol Lett. 2016;11:1745–1749. doi: 10.3892/ol.2016.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50–54. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]