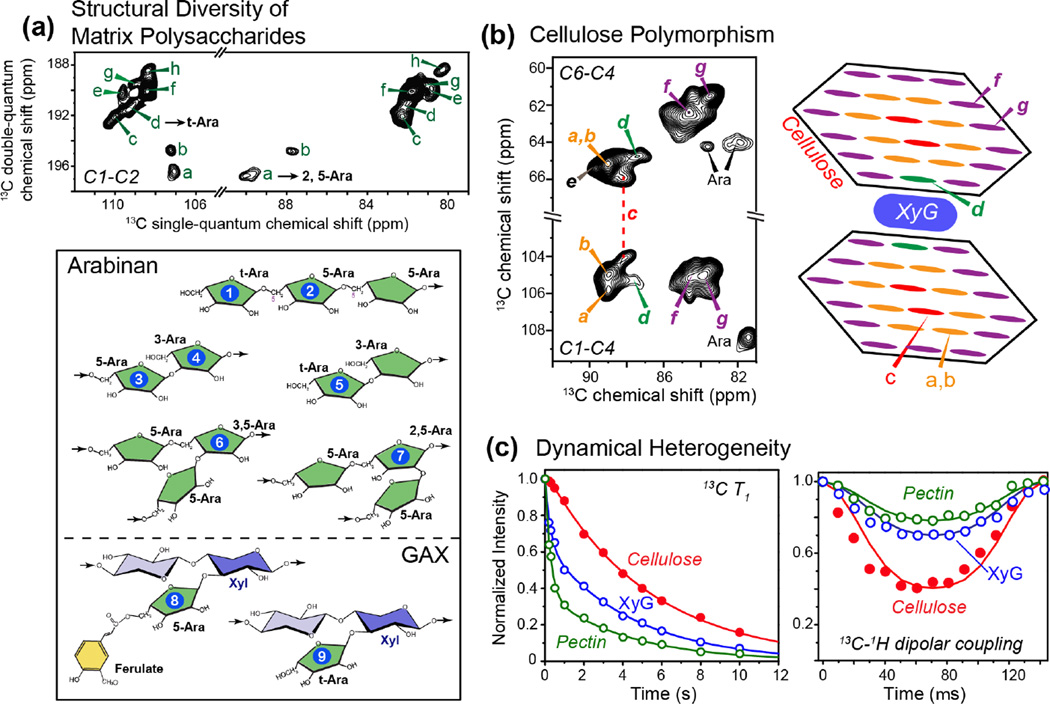
Figure 1.
Structural and dynamical heterogeneity of primary plant cell walls. (a) Structural heterogeneity of arabinose in Brachypodium cell walls [11]. Eight sets of arabinose signals (a–h) are resolved (top), many of which can be assigned to the different linkage types shown at the bottom. (b) Conformational polymorphism of cellulose in Brachypodium cell walls [23]. Seven sets of cellulose signals are partially resolved, with five forms attributed to interior crystalline cellulose (type a–e) and two types to surface cellulose (type f and g). Type-d cellulose can be assigned to XyG-proximal cellulose and is unique to plant cell walls. (c) Representative 13C T1 relaxation data (left) and 13C-1H dipolar coupling data (right) of cellulose (red), hemicellulose XyG (blue) and pectins (green) in Arabidopsis cell walls. Cellulose is mostly rigid while pectins and hemicellulose are partly mobile. The pectin 13C T1 relaxation is double-exponential, with the slow component (~ 5 s) resembling the cellulose 13C T1, indicating that this slow relaxation originates from cellulose-containing pectin segments. The fast-decaying component of ~0.5 s likely results from interfibrillar pectins [15].