Abstract
Understanding how the brain operates requires understanding how large sets of neurons function together. Modern recording technology makes it possible to simultaneously record the activity of hundreds of neurons, and technological developments will soon allow recording of thousands or tens of thousands. As with all experimental techniques, these methods are subject to confounds that complicate the interpretation of such recordings, and could lead to erroneous scientific conclusions. Here, we discuss methods for assessing and improving the quality of data from these techniques, and outline likely future directions in this field.
Introduction
The more powerful an experimental method, the more care must be taken to ensure its correct application. The two leading methods for measuring the activity of many neurons simultaneously – multichannel electrophysiology and population calcium imaging – are benefiting from an exploding range of technical innovations. The increasing complexity of these methods, however, requires increasingly sophisticated approaches to ensure the quality of the data recorded. Quality control is more essential than ever, to ensure the scientific conclusions based on data from these methods are correct, and not a result of experimental artifacts.
In this Review, we discuss some factors that can affect data quality in neuronal population recordings. Careful experiments, of course, are the foundation of high quality data. Experimental design involves inevitable tradeoffs, for example between the number of neurons that can be recorded and the error rates that are acceptable. It is also important that scientists use appropriate data processing techniques to reliably identify individual neurons, and detect when signals are likely to be corrupted. At present – as with many other techniques – the involvement of human operators plays an important role in these quality control procedures, and is currently unavoidable. While effective at catching or correcting data quality problems, input from human operators can also produce biases if not carefully applied.
There can be multiple types of experimental errors in neurophysiology, whose importance depends on the scientific question. False-positive errors – the assignment of spikes to a neuron that did not actually fire them – can lead to invalid conclusions about how information is encoded. False-negative errors – omission of spikes that a neuron genuinely fired – will have a potentially milder consequence of underestimating firing rate and reliability, but only if the errors occur at random: if the errors are themselves correlated with other factors (such as particular patterns of network activity, bursting, or movement), then invalid conclusions could again arise. In population recordings, false positive and false negative errors often arise together, resulting from the incorrect assignment of one cell's spikes to another, from the incorrect merging of multiple cells' spikes together, or from incorrect splitting of a single neuron's spikes into multiple detected cells. These correlated errors can lead to potentially invalid conclusions about population coding and correlation patterns. Furthermore, selection bias (a systematic failure to detect certain types of cells) can give an incorrect picture of how information is encoded at the population level.
Ultimately, a proper understanding of the limitations in current experimental techniques will only be achieved when sufficient “ground truth” data has been collected. In practice, “ground truth” refers to measurement of neural population activity simultaneously with a method such as on-cell electrophysiology, which offers nearly perfect detection of all spikes fired by a single neuron. Such data are presently rare. Nevertheless, the existing ground truth data, together with other approaches such as simulation, has helped the field develop an understanding of the types of confounds that can occur, and methods to identify or correct errors. Careful application of these approaches can help ensure scientific conclusions based on data from population activity measurements are robust.
Extracellular electrophysiology
Extracellular neuronal recordings are typically performed by inserting microelectrodes, insulated everywhere except one or more small recording sites1. The signals from the electrodes are amplified and digitized with a sampling frequency in the range of 20–30KHz, a rate required to resolve extracellular action potentials waveforms ( “spikes”) of duration the order of 1-2 msec. In addition to spike waveforms, the extracellular voltage contains a higher-amplitude, lower frequency “local field potential” signal, which is typically separated from the spike signals by filtering, and used to provide an indirect measure of ongoing global activity patterns.
An electrode with a single recording site can detect the activity of multiple neurons, but to separate the activity of these cells requires appropriate computational analyses. Typically, spikes are detected as crossings of an amplitude threshold, and a waveform is extracted for each spike and temporally realigned to subsample resolution. Because the extracellular spike amplitudes and waveform shapes produced by different neurons at any one point in space can differ2, the firing of individual neurons can be separated by classifying the waveforms into discrete groups, a process known as “spike sorting”3–7. The peak amplitude of the spike decreases with the distance from the neuron to the recording site, and for neurons located closer than ~50 μm from the electrode tip, the spikes are large enough to be detected over background activity and it is often possible to separate them according to their shapes1, 8. For neurons further away, up to about 150–200 micrometers from the electrode tip, spikes can be detected but the difference in their shapes is masked by the noise and they are grouped together as “multiunit activity”. Neurons further away cannot be detected and they contribute to the background activity in the recording.
Why is spike sorting necessary?
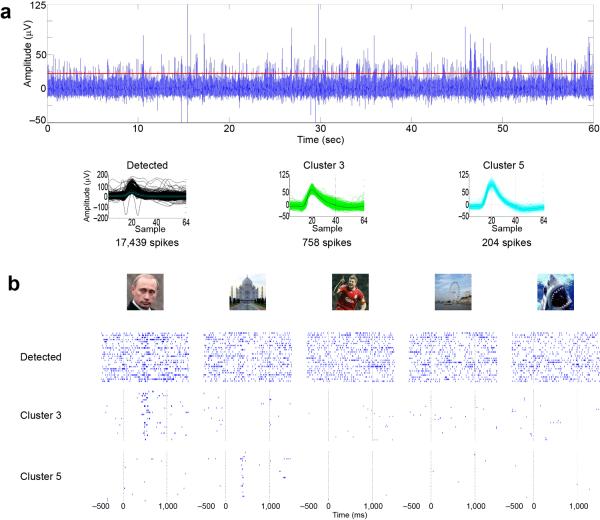
The importance of spike sorting is dramatically illustrated by intracranial recordings made in the human brain. Consider an example recording made with a intracranial microwire electrode, implanted for clinical reasons into the hippocampus of an epilepsy patient, who viewed pictures presented in random order (Figure 1)9, 10. If one were to consider all detected spikes, without spike sorting, no obvious increase over baseline firing rate is visible for any of the stimuli. After separating the neurons based on spike shapes, however, a very different conclusion emerges: the spikes in the original signal reflected the mixed activity of multiple neurons, and these neurons are extremely selective to individual pictures: one neuron responded reliably to a picture of Vladimir Putin, and another responded to a picture of the Taj Majal. The spikes from these two neurons represent only about 4% and 1% of the total number of spikes recorded in this electrode, and this extreme selectivity could not have been detected without spike sorting. In general, the selectivity of single units in the human medial temporal lobe is higher than the selectivity observed for multiunits, where it is not possible to separate the contribution of the different units11. Correct spike sorting is thus essential to understand the neural code employed by this brain circuit. Without it, one would not only miss very sparse responses, but also make the erroneous conclusion that the circuit employs a “dense code”, in which individual units conveyed information about multiple stimuli, rather than a “sparse code” in which single cells are exquisitely tuned for particular stimuli.
Figure 1.
Spike sorting is required to draw valid conclusions in extracellular electrophysiology. (a) Top, extracellular recording from a single microwire electrode in the hippocampus of a patient implanted with intracranial electrodes for clinical reasons. Bottom plots show overlapped all detected spikes (left), and the sorted spikes corresponding to 2 single units (clusters 3 and 5). (b) Responses to 5 pictures presented in an experimental session. Considering all the detected spikes together, no response can be observed in the raster plots. However, a clear response to Vladimir Putin appears when considering only the spikes corresponding to cluster 3, and a response to the Taj Majal appears when considering the spikes corresponding to cluster 5. Time zero corresponds to stimulus onset. For space reasons, only 2 of the 8 identified clusters and only 5 of the 14 presented pictures are shown, but there were no responses for these other clusters and pictures. Adapted from Ref.9.
Electrode design
The physical design of the electrodes used for extracellular electrophysiology makes a great difference to the type of signals recorded. Intracellular recordings represent a “gold standard” in neurophysiology, offering perfect spike detection, as well as the ability to measure and control membrane potentials, estimate synaptic conductances, control the cell's chemical environment, and stain for later anatomical reconstruction. Nevertheless, the difficulty of intracellular recording severely limits its use for large-scale studies. Electrodes placed directly outside the cell (juxtacellular or on-cell recordings) provide a somewhat easier way to obtain perfect isolation, but are still impractical to use at scale. Extracellular recordings – which rely on detection of electric fields tens of microns away from the recorded cell – are more straightforward, but also involve greater errors, which increase with the number of neurons recorded.
The quality of extracellular unit isolation depends on the amplitudes of extracellular spikes relative to background noise. This noise arises from two sources. The first is thermal noise, which increases with the resistance of the recording electrode (e.g. Ref.12), and can be ameliorated by coating with materials such as PEDOT13. The second noise source is the firing of the large number of neurons that are too far from the recording site to produce sortable spikes (>~50 μm), but superimpose to produce “hash” in the same frequency range1, 8.
Different electrode designs offer complementary advantages. Small electrodes (with diameter < 5-10 μm and impedance > 1 MΩ) record from only a few nearby neurons14, but can show excellent unit isolation as they can be positioned very close to the neurons of interest; since spike amplitudes decay rapidly with distance2, 8, this results in large amplitudes relative to background activity. Larger electrodes (diameter several tens of μm) have impedances typically below 1 MΩ, and record the activity of neurons in a larger area. For single sites, a smaller ratio of the amplitudes of nearby neurons to those of the distant cells contributing to the background activity leads to a lower signal-to-noise ratio (SNR). The optimal design of an electrode thus depends on the techniques that are used to process the data: with limited spike sorting algorithms, it might be preferable to have small electrodes with large SNR, whereas with more advanced algorithms and multisite probes, larger, low impedance electrodes can increase the yield of identified neurons15, 16.
An important and understudied question concerns the degree to which electrode insertion damages the neural tissue that is being recorded. Calculations based on the decay of amplitudes with distance suggest that a single tetrode should be able to detect the activity of up to 100 single neurons in hippocampus, but in reality one typically finds an order of magnitude fewer1. Although it is possible that the missing “dark matter” neurons are healthy but not firing, or firing but not identified by current spike sorting algorithms17, insertion of the electrode may also have damaged or killed a substantial fraction of them. Systematic investigation of how electrode geometries, materials, and insertion strategies affect tissue damage would greatly help optimal electrode design.
Multichannel electrophysiology
Understanding complex brain processes requires the analysis of large and simultaneously recorded neuronal populations1, 18–22. Current multielectrode designs allow recording from hundreds of electrodes, and thus hundreds of neurons simultaneously4, 23–27, and these improvements have been matched by increased capabilities of data acquisition systems.
There are two major approaches to the design of multielectrode arrays. The first approach – exemplified by microwire arrays that have been used for animal studies28, or the microfabricated “Utah arrays” that have been implanted in human cortex to enable brain-computer interfaces29 – consists of a large number of single-contact electrodes, with an inter-site distance of at least 100 μm. As any individual neuron can be detected by at most one of the recording sites, data processing for such electrodes (and the corresponding quality concerns) is the same as for single site electrodes. Thus, the recorded population size increases linearly with site count, although without independently movable contacts, many sites may not record neurons. Furthermore, isolation quality does not improve with increasing site count.
The second approach to multielectrode array design is to use dense arrays with an intersite distance of less than 50 micrometers. This approach employs either twisted microwire bundles such as “tetrodes” (4 microwires twisted together30, 31), or micromachined silicon probes1, 32–35. High density probes allow recording the spikes of a single neuron from multiple sites. This improves spike sorting performance, as the spikes of two neurons can frequently look identical on one channel, but differ on others31, 36. With this method, unit isolation quality is expected to scale with site density, as confirmed by spatially subsampling of data from dense electrodes37. Nevertheless, the number of sites achievable is limited by constraints of manufacturing technology, for example shaft diameter (probes that are too thick may damage brain tissue), or the increased noise found with small, high-impedance recording sites. Furthermore, the benefits of high density probes require using spike sorting algorithms that can combine information from different channels.
Spike sorting methods
Spike sorting is more complex for dense arrays than for single contact electrodes. For tetrodes, the traditional method of spike sorting – still commonly applied in many labs – is purely manual cluster cutting. In this procedure, a set of features are computed for each spike, such as the peak amplitude on each channel, or waveform features evaluated by principal component analysis. Using a graphical interface, an operator manually draws boundaries around the resulting “clusters”, which correspond to putative single neurons. The operator is guided in this process by a number of additional tools, such as the computation of auto- and cross-correlograms, which can help identify poorly isolated units by the presence of refractory period violations.
Today, the most commonly employed spike sorting method is “semi-automatic”. With this approach, spike detection and feature extraction proceeds as before, but an automatic cluster analysis algorithm is run on the spike data, and a human operator uses a graphical interface similar to those used for manual sorting to check its output, and correct mistakes the algorithm has made where necessary (some examples of such possible mistakes are described below). Compared to purely manual spike sorting, the semi-automatic approach has two major advantages. The first is time: it takes substantially less human time to check the output of an automatic algorithm, than to perform a fully manual sort. The second advantage is that this approach achieves substantially lower errors than purely manual sorting, as demonstrated using ground-truth tetrode data36. These lower error rates occur because, the optimal boundary between clusters is a high-dimensional surface, which can be found by automatic algorithms but not drawn by human hand using a 2d computer interface36.
The presence of a human operator in the data processing pipeline raises the potential for subjectivity and bias to occur. However, while a fully-automatic spike sorting system would clearly be desirable, it has to date not proved possible to implement algorithms which work robustly in real-world in extracellular recordings. Similar constraints are faced in several other fields of data-intensive biology, such as electron-microscopic connectomics, which also rely on manual operator curation38. Thus, while substantial developments have occurred to reduce the amount of manual operator time required for the sorting process37, 39, fully automatic systems are rarely applied in current practice. Fortunately, analyses comparing the decisions made by multiple expert operators have shown that their corrections of automatic cluster performance tend to be similar17, 37.
Validation of spike sorting
One of the largest problems for the development of spike sorting methods is the paucity of “ground truth” data, in which extracellular arrays are combined with other methods providing unambiguous recording of firing times. While invertebrate and in vitro preparations have provided some data on which to test algorithms40–42, the noise conditions and nonstationarity found in mammalian systems in vivo are substantially more challenging. The difficulty of obtaining ground truth data in vivo has made such data very rare8, 36, 43 (https://crcns.org/data-sets/hc/hc-1). The data that is available, however, suggests that error rates for semi-automatic clustering with tetrodes can be of the order 5-10%, but the error rates of purely manual cluster cutting may be substantially higher36.
In the absence of suitable ground truth data, a number of methods have been used to generate “surrogate” ground truth to validate spike sorting algorithms. One approach consists of performing detailed biophysical simulations of extracellular activity44–46. The computational expense of this method, combined with uncertainty in exactly how to model the challenges of real data has encouraged other authors to try a different method. In this “hybrid” approach, spikes isolated from one recording are digitally added at known times to a second recording, or to a simulation of background activity16, 47, 48. This approach has been used to estimate the errors expected from semi-automatic analysis of high-count array data, again yielding an estimate of errors of the order 5-10%37. An alternative approach relies on measuring the reliability of the spike sorting algorithm under perturbations of the data set49.
Most current methods of spike sorting fail at times of high neuronal synchrony. In the hippocampus, for example, transient events of highly synchronous neural activity known as “sharp waves” are of great interest due to their proposed role in memory. Ground truth data suggests that error rates during sharp waves can be 5 times above average, reaching levels as high as 50% (Ref.36). Such errors could come from two sources. First, synchronous activity might lead to temporally overlapping spikes, which cannot be resolved by traditional spike sorting algorithms; but might be resolvable by newer algorithms based on template matching50–53. Alternatively, ripples might lead to the firing of otherwise silent cells with waveforms too similar to be distinguished accurately; consistent with this possibility, sharp waves are accompanied primarily by an increase in false positive, not false negative errors36.
Common confounds in extracellular electrophysiology
Isolation quality
In even the best quality extracellular recordings, most spikes will come from neurons far from the probe, with amplitudes too low for effective isolation. It is therefore important to identify which clusters correspond to well-isolated single cells, and which represent mixtures of several neurons. The importance of such metrics is underscored by the fact that different operators – although they generally agree on which corrections to make in semi-automatic clustering – can have very different opinions on what constitutes a well-isolated unit37.
To address this issue, several quantitative metrics of cluster quality have been proposed. As all neurons exhibit an absolute refractory period, any cluster that shows a large fraction of inter-spike intervals (ISIs) less than 1-2 milliseconds cannot be a well isolated unit. However, the converse does not apply: an apparently clear refractory period does not imply good quality isolation. Indeed, a cluster that contained the intermixed spikes of two different neurons that fired at separate times (for example hippocampal neurons with non-overlapping place fields) would have a completely clear refractory period. Furthermore, manual examination of autocorrelograms in the presence of bursting can lead to an erroneous impression of a clean refractory period even for very poorly isolated cells36.
A second class of quality metrics measures how well the spikes of one cluster are separated from those of neighboring neurons12, 54–56. It is important to note, however, that there is no single threshold value that objectively defines “good” isolation quality: the criterion must depend on the scientific application. For example, an analysis of the structure of complex spike bursts – which involves a progressive decrease of amplitude as the burst continues (e.g. Refs.57, 58) – required a highly stringent criterion56. Accurate isolation is also critical to study pairwise correlations of spike trains59–61; for example, if the spikes of a single neuron are artificially divided into two clusters, these clusters will show a spurious negative correlation because their spikes will be always separated in time. Estimation of neuronal tuning and selectivity can be also highly sensitive to clustering errors, as demonstrated by the earlier example of human intracranial recordings. By contrast, for brain-machine-interface applications, the exact identity of the neuron generating each spike might not be crucial, and it may be advantageous to use the largest possible number of recording sites in an unsupervised way, even if not sorted at all62, 63. Spike sorting may also be less critical where there is a topographic organization of responses – i.e. when nearby cells tend to fire to similar stimuli – compared to cases when nearby neurons fire to unrelated stimuli, as it has been described in the rodent64 and the human hippocampus9 (Figure 1).
When using an isolation quality metric, how should a scientist decide what threshold value of isolation quality to require for a particular scientific question? A simple method is to consider how the quantity being measured depends on isolation quality. For example, the cross-validated spatial information encoded by putative pyramidal cells of rat CA1 drops substantially for values of isolation distance54 less than 20, but reaches an asymptote above this value (Figure 2). This suggests that a threshold of 20 is suitable for analysis of spatial information coding in these cells.
Figure 2.
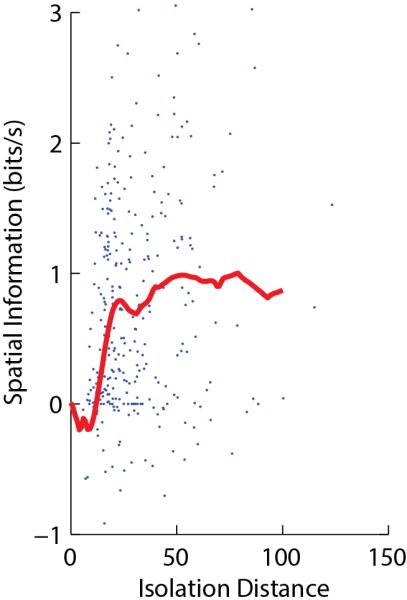
Quantitative measures of unit isolation in extracellular electrophysiology. Each point represents a single neuron recorded in CA1 of a rat exploring an environment, showing isolation distance (a measure of unit isolation quality54) vs. estimated spatial information content (which can be negative as it is computed by cross-validation142). The red curve represents a running median. The curve reaches an asymptote of ~1 bit/s for values of isolation distance greater than ~20, indicating that this is the true average for well-isolated cells. Data reanalyzed from Ref.142, with some points above the top y-axis value truncated for visualization purposes.
Selection bias
Simultaneous intra- and extracellular recordings suggest that there should be approximately 140 single neurons within the radius recordable by a single tetrode in the hippocampus1, 8. However, actual tetrode recordings rarely detect more than a dozen neurons at a time. The reason for this disagreement has been attributed to the presence of silent neurons65, electrical insulation66, damage produced by electrode insertion67 or a potential inability of spike sorting algorithms to deal with large numbers of neurons17.
Both manual and automatic spike sorting methods are likely to be biased against low-rate neurons: if a cell fires only a few spikes, these will not be sufficient to define a cluster and the cell will be missed. Given the preponderance of low-rate cells in brain circuits, a failure to account for this selection bias could lead to incorrect estimation of the firing distribution68. In neocortex, where superficial-layer pyramidal cells fire with lower rates than deep-layer pyramids and fast-spiking (FS) interneurons of all layers69–72, bias towards high-rate neurons has led most population electrophysiology to focused on deep cortical layers.
Historically, single neuron recordings have been performed by advancing the electrodes until neural activity is detected73. This method can lead to a different form of selection bias: not only will recordings be made primarily from high-rate neurons (perhaps again leading to a bias toward FS interneurons), but also from cells responding to the specific stimuli or conditions present at the time of the recording. Without care, this could lead to a “confirmation bias”: an investigator would find an over-abundance of neurons that respond precisely to the stimulus or condition being investigated.
Sampling bias in extracellular electrophysiology can be ameliorated by performing non-stop chronic recordings, using fixed electrodes, over very long time periods: recording 24 hours/day for days or weeks can lead to sufficient spike numbers to define clusters even for cells of very low firing rate39.
Operator bias
Because spike sorting has a manual curation step, the possibility of subconscious operator biases must be very carefully excluded. As with many other procedures, when comparing recordings of subjects in different conditions, it is essential that the operator performs manual curation blinded to the condition of each recording. This is particularly important when analyzing quantities such as stability of firing patterns, which can be easily altered by the manual curation step74.
Another important concern regards the use of the neuron's firing correlates (e.g. sensory receptive fields or place fields) in the spike sorting process. While observing a similar firing pattern in two clusters does make it more likely that they represent the same cell, the use of receptive field information during clustering may bias results. Indeed, if neurons genuinely show receptive field plasticity, this will be underestimated if response stability is taken as a criterion for good isolation.
Electrode and waveform drift
Errors in spike sorting are of two types: the spikes of different cells can be erroneously merged together, or the spikes of a single cell can be erroneously separated into two or more clusters (“overclustering”), which often occurs when the spike waveforms of a particular neuron varies during the course of the experiment.
Waveform variability can occur for multiple reasons. The most common reason is “electrode drift”: the physical movement of the electrode relative to the brain. Due to the highly localized electric fields neurons produce2, 8, 16, even a few microns' movement is enough to cause substantial variability in spike amplitudes. This variability is largest for high amplitude spikes, consistent with the sharply-peaked structure of extracellular electric fields. This problem is particularly severe in acute recordings, in which the electrode is fixed not to the skull, but to an external manipulator, meaning that small movements of the head, or relaxation of the brain after compression caused by probe insertion, will cause a movement of the cells relative to the electrodes. Physical electrode drifts are less serious in chronic recordings, where stable recordings have been observed for periods of days or weeks39, 75.
Not all waveform variability is caused by physical movement. Extracellular spike amplitudes decrease during the course of complex-spike bursts, and also decrease after prolonged firing, even without bursting56, 58. Moreover, extracellular waveform shapes can depend on cellular factors such as dendritic action potential backpropagation or electrogenesis, which can vary with firing history, inhibition and neuromodulation76, 77. The difficulty of quantitatively modeling these phenomena is one reason fully automatic spike sorting has so far proved challenging; nevertheless, the fact that these effects can be caught during manual curation suggests that automatic systems may also be eventually possible.
Outlook
While today's silicon probes have at most a few hundred channels, probes with thousands of channels are currently under development. These probes will raise new challenges for data processing and quality control, the most important concerning manual spike sorting. Purely manual sorting is clearly impossible for this size of data, and even curation of semi-automatic sorting will become a serious burden. This burden can be dramatically reduced by the development of algorithms that minimize operator time, by directing attention to only those decisions that cannot be made automatically. Fully automatic spike sorting not only becomes more desirable with very high count probes, but it may also become more achievable. Electrode drift and the consequent spike shape changes presents one of the biggest barriers to fully automatic sorting; it is possible that large dense probes might sample the extracellular electric fields with enough spatial resolution to allow drift to be tracked and corrected in software.
A second challenge for spike sorting regards the long-term tracking of clusters, to study plasticity, for example, during learning experiments. With a few exceptions, scientists have been hesitant to use extracellular electrophysiology to study long-term plasticity of firing properties, as clusters may appear, disappear, merge or separate78–80, and it is critical not to confuse changes in tuning of neuronal populations with changes in the recording conditions or small electrode movements. Three techniques may ameliorate these problems. The first is the gradual refinement of chronic recording methods, which can now ensure high stability of many cells over multiple days or even weeks of recordings39, 75, 81. The second is the development of quantitative methods for unbiased assessment of cluster similarity, that may help identify neurons across multiple days78, 82–84. The third is the use of 24-hour recording39 which reduces the problem to a much easier one of tracking slow continuous changes, than tracking across sudden jumps between recording sessions.
Finally, and most important, there is a need for research into fundamental questions underpinning extracellular array recording. What electrode geometries, surface contact diameters, impedances, and materials provide the best data quality while avoiding tissue damage? How do these properties interact with the choice of sorting algorithms? The most critical experiments to solve this problem involve collection of ground truth data, to quantify the performance of different electrodes and sorting methods without relying on simulations.
Population recording via calcium imaging
Calcium imaging is a complementary technique for measuring the activity of neuronal populations. Depolarization during action potentials opens voltage-gated Ca2+ channels and results in a transient increase in intracellular [Ca2+], which can be detected optically using fluorescent reporters: calcium-sensitive dyes or proteins. Calcium imaging can be used to infer patterns of spiking activity across hundreds to thousands of identified cells in vivo85, 86. Like any measurement, however, it demands careful application and analysis.
Calcium signals are correlated with neuronal spiking, but are an indirect reflection of it, and biophysical variations make the precise relationship between calcium signals and spiking variable87. Calcium reporters can themselves limit the precision of spike inference: although synthetic calcium dyes can be used in a linear regime, they still exhibit nonlinear features including saturation; and genetically encoded calcium indicator (GECI) proteins are highly nonlinear due to cooperative Ca2+ binding88.
The temporal resolution of the calcium signal is limited. Indicator kinetics are relatively slow (e.g., rise times of ~10 ms for single action potentials measured with synthetic dyes, and >50 ms for many GECIs). Furthermore, imaging methods that scan over space trade off recordable population size against sampling time. Recordings from large neuronal populations often require timesteps on the order of tens to hundreds of ms (Ref.86).
As with electrophysiology, the ultimate check on recording quality is ground truth, typically obtained by simultaneous on-cell patch recordings. Such recordings are feasible with some, but not all calcium imaging instrumentation. However, even in the absence of ground truth, good experimental design and rigorous analysis can improve data quality. Below, we discuss these considerations. Our discussion primarily relates to two-photon laser scanning microscopy, but several points are relevant to one-photon imaging, including wide-field imaging through GRIN lenses89, and light-sheet imaging in transparent specimens90.
Experimental design and measurement noise
As with any technique, instrumentation, preparation, and recording parameters must be tailored to the demands of the specific scientific application. Some experiments require detecting single action potentials and/or resolving precise spike counts in each neuron with minimal uncertainty. For others, detecting qualitative increases and decreases in spike rate suffices. Experimental design is critical because most optimizations involve tradeoffs. For example, high zoom (many pixels per neuron) and high frame rate can provide faithful estimates of spiking based on calcium signals91, but also limit the number of neurons that can be sampled in each imaging frame. Low frame rate acquisition can be sufficient to map population responses when the stimulus changes slower than the acquisition rate: for example, Ohki and colleagues92 mapped the orientation-tuned responses of hundreds of neurons at 0.61 frames / s, with a stimulus that changed at 0.0625 Hz. By contrast, Dombeck and colleagues93 acquired 15.6 frames/s over smaller fields-of-view, to capture the activity of hippocampal place fields with sub-second resolution.
Imaging quality depends primarily on recording a sufficient number of photons per pixel. Photon emission exhibits Poisson-like variability (“shot noise”), with SNR scaling as the square root of the photon count94. Optimizing quality thus requires tuning parameters such as laser power and scan configuration, which includes parameters such as pixel dwell time, field-of-view, pixels per neuron, subject to the limitation that the overly-high laser power can cause tissue damage95. Optimized scan patterns96–98 can target sampling to individual neurons, but lack the spatial coverage required for posthoc motion correction. Photons per pixel can be estimated from parameters including the gain and offset of the photomultiplier tube (http://labrigger.com/blog/2010/07/30/measuring-the-gain-of-your-imaging-system/). Tradeoffs between photons per pixel, numbers of pixels, and SNR can quantified using signal detection theory, and compared and optimized for specific experiments99, 100,101. For example, GCaMP6s imaging can provide single AP detection with nearly 100% detection of all spikes when imaged at a high frame rate over a small field-of-view (30 μm × 30 μm at 60 frames/s)91. However, larger field-of-view, population-level imaging (265 μm × 265 μm at 59.1 frames/s) yields spike rate estimates that correlate with the true spike rate at an average level of ~ 0.5 (Pearson's R, using a 50 ms spike rate binning window)87. Quality is also affected by indicator properties and labeling intensity88, 94. Typically, however, these quantities are not measured for in vivo preparations, and parameters yielding high SNR data are identified by trial-and-error (e.g., dye concentration, viral vector titer, number of days after transfection to image).
Signal contamination
In densely labeled tissue, structures adjacent to cell bodies can contribute contaminating signals102. In areas such as neocortex, somata are distributed sparsely enough to make contamination from adjacent cell bodies rare103, permitting moderate-resolution imaging systems to accurately measure cellular-resolution dynamics92, 103, 104. However, in structures with densely packed cell bodies, such as the hippocampus or cerebellar granular layer, cell-to-cell contamination cannot be ignored.
Calcium imaging of large populations typically leads to contamination from signals arising in the neuropil: the axons and dendrites of nearby cells, whose activity produces a substantial contamination of the signal recorded at any cell soma91, 105, 106. Even with two-photon microscopy, axial resolution is often limited to several microns or more, although in-plane (lateral) resolution can be sub-micron (Fig. 3a). Furthermore, even high resolution (i.e. high numerical aperture) imaging systems are precluded from realizing their full resolution in practice, due to optical aberrations caused by brain tissue and the loss of marginal rays when imaging deep107, 108. Imaging system aberrations can also limit resolution, particularly when imaging outside of the very center of the field-of-view109,110. GRIN lenses, used to access deep structures, also suffer from significant aberrations111,112. Adaptive optics can compensate for aberrations of the focused excitation light113, but the emitted fluorescence photons are still subjected to scattering and aberrations in the tissue and optical systems. Neuropil contamination can be somewhat mitigated by expressing GECIs in sparsely, for example using transgenic mice with Thy1 promoters, or conditional viral strategies91, 114–116. However, neuropil contamination represents a serious concerns for calcium imaging. The neuropil signal reflects the summed activity of a large number of neurons, the majority of which will typically be located close to the imaged region, and can thus itself be tuned for similar stimuli or behavioral variables117. If not accounted for, neuropil contamination can lead to spurious conclusions about neuronal encoding. It is therefore essential that this contamination be understood and, to the extent possible, removed during analysis. Approaches will be discussed in “data processing”, below.
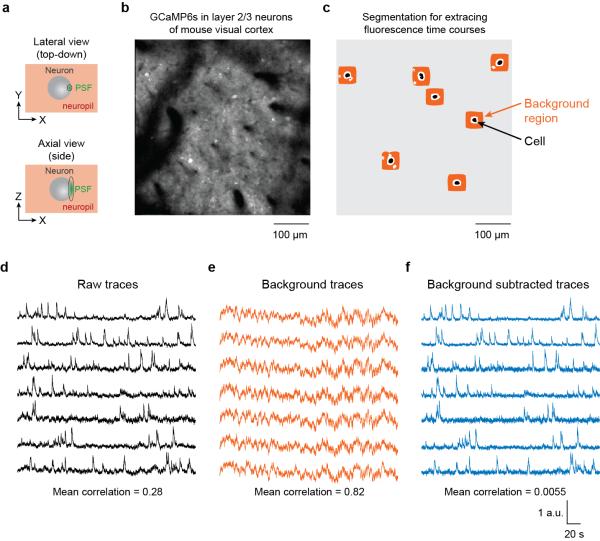
Figure 3.
Subtracting neuropil contamination from raw fluorescence time courses. (a) In two-photon imaging, the point spread function (PSF) is elongated in the axial dimension even in high numerical aperture systems. Pixels within the borders of cell bodies still contain signals from the surrounding neuropil. (b) The GECI GCaMP6s was expressed in mouse visual cortex neurons, resulting in brightly labeled cell bodies and neuropil. (c) Binary masks for cell body ROIs (black) were identified semi-automatically and neuropil regions were algorithmically constructed (avoiding pixels belonging to other potential cell bodies or black regions). (d) Raw traces for the fluorescence time courses of the selected cells. (e) Fluorescence time courses for the background regions for each selected cell. Note the high temporal correlation. (f) Fluorescence time courses after background subtraction. Note the reduced mean correlation141. All traces have been scaled to the same maximum height to better exhibit details in the time courses. Figure credit: J. N. Stirman, Y. Yu, S. L. Smith
Motion artifacts
When imaging in living subjects, heartbeat, breathing, and motor behavior can all contribute significant movement artifacts. While heartbeat-associated movements in the brain are typically on the order of 1 μm or less118, breathing and motor behavior can cause larger movements over 10 μm119, 120. Movement amplitudes vary by brain area and can be reduced by appropriate surgical preparation121. During movements, cells can change their pixel location in the imaging plane (XY), or in and out of the plane of focus (Z). If temporal resolution is high enough to make frame-to-frame XY movement small, movement can be corrected through image registration, e.g. global cross-correlation or line-by-line alignment117, 122, 123. Large and faster XY movements may require more complex model-based algorithms119, 124. Motion in Z is more difficult to correct, since the neighboring planes are typically not recorded. Generally, point spread functions (PSFs) are extended in Z, making motion artifacts due to small Z movements less problematic125, 126. In cases with large Z movement, multiplane imaging127 and online motion correction120, 128 can help minimize artifacts.
Data processing
Before calcium data can be used for scientific analysis, it requires preprocessing, typically involving: image alignment/motion correction; segmentation to find regions of interest (ROIs) corresponding to imaged cells; time course extraction; neuropil compensation; and, optionally, spike train estimation. In some analysis algorithms, multiple steps are performed in concert.
Segmentation and time course extraction
The simplest way to define ROIs corresponding to individual neurons is with binary masks, which can be drawn manually or with varying degrees of automation85. With binary masks, neuropil contamination must be estimated and subtracted out in a separate step, and can be estimated using either an average of the neuropil signal surrounding the cell of interest129 (Fig. 3b–f), or by subtracting the first principal component of the contamination130, 131.
More ambitious approaches describe each pixel's calcium signal as a superposition of signals from one or more cell bodies or processes, neuropil, and potentially other sources (e.g., instrumental noise). This is typically framed as a matrix factorization problem, where the spatial components (neuronal ROIs) and temporal components (neuronal time courses) are simultaneously learned. As with spike-sorting, the results of such automatic algorithms must be verified on a case-by-case basis. For example, methods based on PCA/ICA131, 132 can identify negative temporal signals, which cannot represent neuronal time courses; these methods can also produce ROIs corresponding multiple neurons or dendritic regions, since the algorithm has no prior information about the spatial extent of signal sources. Constrained nonnegative matrix factorization, optimized for the particular characteristics of calcium imaging data133, 134 provides promising results even in the presence of signal crosstalk106, while a new combined clustering and factorization method can accurately process over 10,000 cells in approximately real time135. As matrix factorization methods separate temporally distinct patterns of activity, they may fail on neighboring cells that exhibit highly synchronous activity; a supervised learning approach might avoid this shortcoming96. Furthermore, any activity-dependent algorithm will be biased towards active cells; labeling cell nuclei with static (not calcium-dependent) fluorescent proteins can help identify cell bodies independent of activity117. Similarly, “dictionary” methods have been developed to detect cells based on average/resting fluorescence activity, thus finding a large number of inactive cells136.
Manually verifying the results of automatic algorithms is time-consuming, but some simple automatic techniques can catch many artifacts. The zero-lag cross-correlation of ROI pairs can identify contamination of one cell's signal by another. These correlations should usually be close to zero, rarely above 0.5, and values > 0.8 typically indicate contamination. Minor contamination can result in smaller increases in correlation values, and so time courses from nearby cells should be checked particularly closely. Correlation due to contamination should be more stable in time than correlation due to neuronal firing, so rolling correlation analysis (with width substantially above that of a fluorescence transient) can help identify unusually stable correlations due to contamination. Excluding individual highly-contaminated pixels will result in less contaminated ROIs. As genuine neuronal correlations can be affected by brain state, this analysis is best performed on data obtained under conditions of relative desynchronization (e.g., stimulus-evoked activity rather than spontaneous activity under anesthesia). Finally, correlating the time-series of individual pixels to stimulus and behavioral variables can help diagnose artifacts.
Evaluation of segmentation approaches would benefit from a systematic comparison of algorithms against ground-truth. Ground truth for segmentation can be hard to obtain, or even define, but options include hand annotation, or co-expressing anatomical markers (e.g. fluorescent proteins that are confined to the nucleus) that report the presence or absence of a neuron soma at a location. The Neurofinder challenge provides several ground truth datasets and a web application for comparing algorithm results (http://neurofinder.codeneuro.org/), and ground truth data is also available at https://crcns.org/data-sets/methods/cai-1. Surprisingly, nearly all segmentation algorithms thus far have taken an unsupervised approach – trying to infer neurons directly from data, rather than by training a supervised model on existing annotations, as is common in object recognition, behavioral classification, and anatomical segmentation. Especially with the availability of annotated data, supervised methods could be a fruitful avenue of exploration.
Spike inference
The calcium signal is only an indirect reflection of spiking. Many analysis approaches aim to derive from the calcium fluorescence time course of each neuron an estimate of firing rates or exact spike times.
The calcium signal can be approximated as a temporally filtered version of the spike train. Although the decay kinetics of this filter are typically slow (half-decay times are hundreds to thousands of ms), its rise kinetics can be fast (times-to-peak are tens of ms). The simplest approach is thus deconvolution with an estimated unitary response, such as an instantaneous rise and exponential decay137, 138. In practice, deconvolution performance is limited by several factors. The relationship between calcium signals and spiking is complex, nonlinear, and varies across neurons, especially during spike bursts. With GCaMP6f, for example, spike pairs can exhibit fluorescence transients whose size depends on inter-spike interval (personal communication from D. DiGregorio and S. Wang). As with segmentation, supervised learning methods using systematic ground truth data are a promising alternative to unsupervised deconvolution87. With present technology, estimates of spike times from calcium imaging should always be treated as approximations, though this uncertainty can be propagated through stages of analysis (Fig. 4)133.
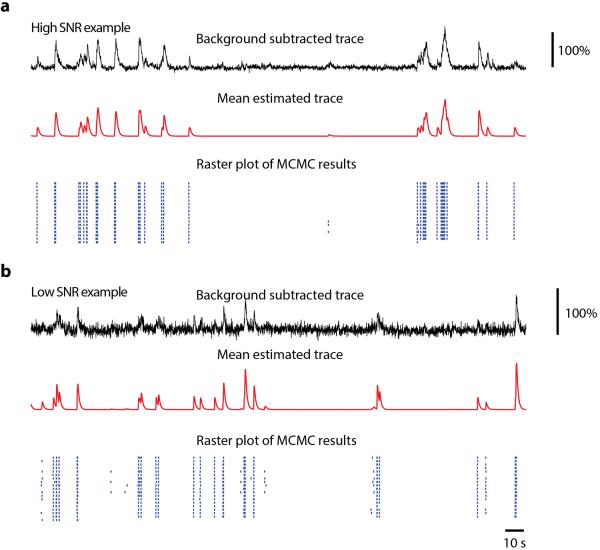
Figure 4.
Probabilistic estimation of spike times from calcium signals. A model of calcium signal generation is inverted to yield a probability distribution of spike trains (blue raster), given a single observed calcium trace (black). (a), high signal-to-noise data leads to a highly certain estimate of spiking. (b), low signal-to-noise leads to an uncertain estimate, illustrated by the raster showing different spike trains that could have generated the calcium signal. Subsequent analysis can be performed using the entire spike train distribution. Figure credit: J. N. Stirman, Y. Yu, S. L. Smith.
Estimation of spike times is not necessary for many scientific questions. When neurons fire sparsely, for example, neuronal responses can be characterized by how the calcium response itself depends on stimulus or behavioral-related factors. The results of such analyses will not be numerically identical to analyses computed from actual counts (for example when computing correlations among neurons), but if interpreted correctly, this can avoid biases introduced by explicit spike estimation. This concept is explicitly formulated in hierarchical models that describe the calcium response as a function of spike times, which are in turn a function of the stimulus and/or behavior. The parameters of such an “encoding” function can be estimated directly without explicitly estimating spike times, instead treating them as latent factors (e.g. Refs.139, 140). Finally, spike detection can be simply viewed as a nonlinear denoising step to remove spurious low-amplitude signals, rather than an explicit estimator of spike times; in this view, procedures based on template matching, which are more flexible and less computationally expensive, may be appealing117.
Ground truth from electrophysiology
Experiments verifying 2-photon calcium imaging with ground truth from simultaneous on-cell electrophysiology have indicated encouraging results. It is important to match the imaging parameters between ground truth data sets and actual experiments: highly zoomed-in imaging offers a “best-case scenario” about how faithfully an indicator might report spiking, but may vary markedly from those obtained when imaging is zoomed-out to increase the number of neurons imaged (which also decreases the SNR of recorded cells). Single action potentials can be resolved >80% of the time in optimized systems, and multi-spike bursts are even more reliably detected91, 141. However, estimates of the number of spikes in multi-spike bursts is typically imprecise, and in practice larger fields-of-view decrease SNR and lead to overall correlation coefficients between 0.1 and 0.5 (Ref.87), though in some studies this has ranged up to 0.8 (Refs.118, 141).
In some cases, it is impossible to perform calibration electrophysiology experiments. For example, the small working distance of GRIN lenses largely precludes correlative electrophysiology. Similarly, with air immersion objectives, simultaneous electrophysiology would so perturb the optical setup as to yield it largely irrelevant. In these cases – where ground truth is unobtainable – experiments should be designed to be insensitive to the expected imprecision of spike inference. To bracket the precision of the estimated spike trains, experimenters can compare their data to electrophysiological recordings under similar circumstances. For example, data from extracellular recordings in mouse visual cortex can provide a baseline for calcium imaging studies, setting both the expected spontaneous firing rates and expected distribution of maximal firing rates in mouse visual cortex in response to drifting grating visual stimuli71, bearing in mind the selection bias in electrophysiology towards active cells.
Summary
Calcium imaging and extracellular electrophysiology can both provide high fidelity readout of neuronal population activity. They have complementary advantages: electrophysiology allows detection of single action potentials with submillisecond timing, in deep structures and multiple brain areas; calcium imaging can provide a comprehensive and less biased view of a local population, and interfaces easily with the genetic toolkit required to identify neurons based on cell type or connectivity.
Both methods are subject to experimental confounds, most notably spike sorting errors for electrophysiology, and signal contamination for calcium imaging. While these confounds cannot be avoided, they can be mitigated through careful experimental design. Furthermore, a detailed understanding of causes and consequences of these confounds makes erroneous scientific conclusions unlikely if they are used carefully. When objective measures of data quality exist, it is important that these are used and documented together with the scientific conclusions drawn. To gain a truly quantitative understanding of the error rates likely to occur in population recordings, however, it is essential that substantial further effort be put into collecting “ground truth” data calibrating these techniques against reliable measures of neural activity.
Acknowledgements
We thank N. Steinmetz and M. Pachitariu for comments on the manuscript. K.D.H is supported by the Wellcome Trust (95668, 95669, 100154), EPSRC (K015141, I005102), MRC, and Simons foundation (SCGB 325512). S.L.S. is supported by the Human Frontier Science Program (CDA00063/2012, RGP0027/2016;), National Science Foundation (1450824), Whitehall Foundation, Klingenstein Foundation, McKnight Foundation, Simons Foundation (SCGB 325407SS), and the National Institutes of Health (R01NS091335, R01EY024294). R.Q.Q is supported by the Human Frontiers Science Program (RGP0015/2013). The authors declare no competing financial interests.
References
- 1.Buzsaki G. Large-scale recording of neuronal ensembles. Nat Neurosci. 2004;7:446–451. doi: 10.1038/nn1233. [DOI] [PubMed] [Google Scholar]
- 2.Gold C, Henze DA, Koch C, Buzsaki G. On the origin of the extracellular action potential waveform: A modeling study. J Neurophysiol. 2006;95:3113–3128. doi: 10.1152/jn.00979.2005. [DOI] [PubMed] [Google Scholar]
- 3.Quian Quiroga R. Quick guide: Spike sorting. Current Biology. 2012;22:R45–R46. doi: 10.1016/j.cub.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Berenyi A, et al. Large-scale, high-density (up to 512 channels) recording of local circuits in behaving animals. Journal of Neurophysiology. 2014;111:1132–1149. doi: 10.1152/jn.00785.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewicki MS. A review of methods for spike sorting: the detection and classification of neural action potentials. Network. 1998;9:R53–R78. [PubMed] [Google Scholar]
- 6.Rey HG, Pedreira C, Quian Quiroga R. Past, present and future of spike sorting techniques. Brain Res Bull. 2015;119:106–117. doi: 10.1016/j.brainresbull.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Einevoll GT, Franke F, Hagen E, Pouzat C, Harris KD. Towards reliable spike-train recordings from thousands of neurons with multielectrodes. Curr Opin Neurobiol. 2012;22:11–17. doi: 10.1016/j.conb.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henze DA, et al. Intracellular features predicted by extracellular recordings in the hippocampus In vivo. J.Neurophysiol. 2000;84:390–400. doi: 10.1152/jn.2000.84.1.390. [DOI] [PubMed] [Google Scholar]
- 9.Rey H, et al. Single cell recordings in the human medial temporal lobe. Journal of Anatomy. 2015;227:394–408. doi: 10.1111/joa.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quian Quiroga R. Concept cells: The building blocks of declarative memory functions. Nature Reviews Neuroscience. 2012;13:587–597. doi: 10.1038/nrn3251. [DOI] [PubMed] [Google Scholar]
- 11.Quian Quiroga R, Reddy L, Koch C, Fried I. Decoding Visual Inputs From Multiple Neurons in the Human Temporal Lobe. J Neurophysiol. 2007;98:1997–2007. doi: 10.1152/jn.00125.2007. [DOI] [PubMed] [Google Scholar]
- 12.Hill DN, Mehta SB, Kleinfeld D. Quality metrics to accompany spike sorting of extracellular signals. J Neurosci. 2011;31:8699–8705. doi: 10.1523/JNEUROSCI.0971-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig KA, Uram JD, Yang J, Martin DC, Kipke DR. Chronic neural recordings using silicon microelectrode arrays electrochemically deposited with a poly(3,4-ethylenedioxythiophene) (PEDOT) film. J Neural Eng. 2006;3:59–70. doi: 10.1088/1741-2560/3/1/007. [DOI] [PubMed] [Google Scholar]
- 14.Gerstein GL, Clarke WA. Simultaneous Studies of Firing Patterns in Several Neurons. Science. 1964;143:1325–1327. [PubMed] [Google Scholar]
- 15.Robinson DA. The electrical properties of metal microelectrodes. Proceedings IEEE. 1968;56:1065–1071. [Google Scholar]
- 16.Camunas-Mesa L, Quian Quiroga R. A detailed and fast model of extracellular recordings. Neural Computation. 2013;25:1191–1212. doi: 10.1162/NECO_a_00433. [DOI] [PubMed] [Google Scholar]
- 17.Pedreira C, Martinez J, Ison M, Quian Quiroga R. How many neurons can we see with current spike sorting algorithms? Journal of Neuroscience Methods. 2012;211:58–65. doi: 10.1016/j.jneumeth.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quian Quiroga R, Panzeri S. Principles of neural coding. Taylor & Francis/CRC Press; Boca Raton: 2013. [Google Scholar]
- 19.Buzsaki G. Neural Syntax: Cell Assemblies, Synapsembles, and Readers. Neuron. 2010;68:362–395. doi: 10.1016/j.neuron.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown EN, Kass RE, Mitra PP. Multiple neural spike train data analysis: state-of-the-art and future challenges. Nat Neurosci. 2004;7 doi: 10.1038/nn1228. [DOI] [PubMed] [Google Scholar]
- 21.Quian Quiroga R, Panzeri S. Extracting information from neural populations: Information theory and decodign approaches. Nature Reviews Neuroscience. 2009;10:173–185. doi: 10.1038/nrn2578. [DOI] [PubMed] [Google Scholar]
- 22.Harris KD. Neural signatures of cell assembly organization. Nat.Rev.Neurosci. 2005;6:399–407. doi: 10.1038/nrn1669. [DOI] [PubMed] [Google Scholar]
- 23.Spira ME, Hai A. Multi-electrode array technologies for neuroscience and cardiology. Nature Nanotechnology. 2013;8:83–94. doi: 10.1038/nnano.2012.265. [DOI] [PubMed] [Google Scholar]
- 24.Lambacher A, et al. Identifying firing mammalian neurons in networks with high-resolution multitransistor array (MTA) Applied Physics A. 2011;102:1–11. [Google Scholar]
- 25.Frey U, Egert U, Heer F, Hafizovic S, Hierlemann A. Microelectronic system for high-resolution mapping of extracellular electric fields applied to brain slices. Biosensors and bioelectronics. 2009;24:2191–2198. doi: 10.1016/j.bios.2008.11.028. [DOI] [PubMed] [Google Scholar]
- 26.Litke A, et al. What does the eye tell the brain?: development of a system fo the large-scale recording of retinal output activity. IEEE Trans. Nucl. Sci. 2004;51:1434–1440. [Google Scholar]
- 27.Lopez CM, et al. 2016 IEEE International Solid-State Circuits Conference (ISSCC); IEEE; 2016. pp. 392–393. [Google Scholar]
- 28.Nicolelis MA, Ghazanfar AA, Faggin BM, Votaw S, Oliveira LM. Reconstructing the engram: simultaneous, multisite, many single neuron recordings. Neuron. 1997;18:529–537. doi: 10.1016/s0896-6273(00)80295-0. [DOI] [PubMed] [Google Scholar]
- 29.Hochberg LR, et al. Neuronal ensemble control of prosthetic devices by a human with tetraplegia. Nature. 2006;442:164–171. doi: 10.1038/nature04970. [DOI] [PubMed] [Google Scholar]
- 30.McNaughton BL, O'Keefe J, Barnes CA. The stereotrode: a new technique for simultaneous isolation of several single units in the central nervous system from multiple unit records. Journal of Neuroscience Methods. 1993;8:391–397. doi: 10.1016/0165-0270(83)90097-3. [DOI] [PubMed] [Google Scholar]
- 31.Gray CM, Maldonado PE, Wilson M, McNaughton B. Tetrodes markedly improve the reliability and yield of multiple single-unit isolation from multi-unit recordings in cat striate cortex. J Neurosci Methods. 1995;63:43–54. doi: 10.1016/0165-0270(95)00085-2. [DOI] [PubMed] [Google Scholar]
- 32.Blanche TJ, Spacek MA, Hetke JF, Swindale NV. Polytrodes: high-density silicon electrode arrays for large-scale multiunit recording. J Neurophysiol. 2005;93:2987–3000. doi: 10.1152/jn.01023.2004. [DOI] [PubMed] [Google Scholar]
- 33.Drake KL, Wise KD, Farraye J, Anderson DJ, BeMent SL. IEEE Transaction on Biomedical Engineering. 1988;Vol. 35:719–732. doi: 10.1109/10.7273. [DOI] [PubMed] [Google Scholar]
- 34.Bai Q, Wise K. Single-unit neural recording with active microelectrode arrays. IEEE Transaction on Biomedical Engineering. 2001;48:911–920. doi: 10.1109/10.936367. [DOI] [PubMed] [Google Scholar]
- 35.Csicsvari J, et al. Massively parallel recording of unit and local field potentials with silicon-based electrodes. J. Neurophysiol. 2003;90:1314–1323. doi: 10.1152/jn.00116.2003. [DOI] [PubMed] [Google Scholar]
- 36.Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsaki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J.Neurophysiol. 2000;84:401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- 37.Rossant C, et al. Spike sorting for large, dense electrode arrays. Nat Neurosci. 2016;19:634–641. doi: 10.1038/nn.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 39.Dhawale AK, et al. Automated long-term recording and analysis of neural activity in behaving animals. bioRxiv. 2015:033266. doi: 10.7554/eLife.27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anastassiou CA, Perin R, Buzsaki G, Markram H, Koch C. Cell type- and activity-dependent extracellular correlates of intracellular spiking. J Neurophysiol. 2015;114:608–623. doi: 10.1152/jn.00628.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wehr M, Pezaris J, Sahani M. Simultaneous paired intracellular and tetrode recordings for spike sorting algorithms. Neurocomputing. 1999;26:1061–1068. [Google Scholar]
- 42.Deleschuse M, Pouzat C. Efficient spike-sorting of multi-state neurons using inter-spike intervals information. Journal of Neuroscience Methods. 2006;150:16–29. doi: 10.1016/j.jneumeth.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 43.Neto JP, et al. Validating silicon polytrodes with paired juxtacellular recordings: method and dataset. bioRxiv. 2016:037937. doi: 10.1152/jn.00103.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linden H, et al. LFPy: a tool for biophysical simulation of extracellular potentials generated by detailed model neurons. Frontiers in neuroinformatics. 2014;7:41. doi: 10.3389/fninf.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menne K, Folkers A, Malina T, Maex R, Hofmann U. Test of spike-sorting algorithms on the basis of simulated network data. Neurocomputing. 2002;44–46:1119–1126. [Google Scholar]
- 46.Petersen KE, Einevoll GT. Amplitude variability and unit isolation from multi-unit recordings in cat striate cortex. Biophysics Journal. 2008;94:784–802. [Google Scholar]
- 47.Martinez J, Pedreira C, Ison M, Quian Quiroga R. Realistic simulation of extracellular recordings. Journal of Neuroscience Methods. 2009;184:285–293. doi: 10.1016/j.jneumeth.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 48.Navajas J, et al. Minimum Requirements for Accurate and Efficient Real-Time On-Chip Spike Sorting. Journal of Neuroscience Methods. 2014;230:51–64. doi: 10.1016/j.jneumeth.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnett AH, Magland JF, Greengard LF. Validation of neural spike sorting algorithms without ground-truth information. J Neurosci Methods. 2016;264:65–77. doi: 10.1016/j.jneumeth.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 50.Franke F, Natora M, Boucsein C, Munk MH, Obermayer K. An online spike detection and spike classification algorithm capable of instantaneous resolution of overlapping spikes. J Comput Neurosci. 2010;29:127–148. doi: 10.1007/s10827-009-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pillow JW, Shlens J, Chichilnisky EJ, Simoncelli EP. A model-based spike sorting algorithm for removing correlation artifacts in multi-neuron recordings. PLoS One. 2013;8:e62123. doi: 10.1371/journal.pone.0062123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ekanadham C, Tranchina D, Simoncelli EP. A unified framework and method for automatic neural spike identification. J Neurosci Methods. 2014;222:47–55. doi: 10.1016/j.jneumeth.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pachitariu M, Steinmetz NA, Kadir SN, Carandini M, Harris KD. Kilosort: realtime spike-sorting for extracellular electrophysiology with hundreds of channels. BioRxiv. 2016:061481. [Google Scholar]
- 54.Schmitzer-Torbert N, Jackson J, Henze D, Harris K, Redish AD. Quantitative measures of cluster quality for use in extracellular recordings. Neuroscience. 2005;131:1–11. doi: 10.1016/j.neuroscience.2004.09.066. [DOI] [PubMed] [Google Scholar]
- 55.Pouzat C, Mazor O, Laurent G. Using noise signature to optimize spike-sorting and to assess neuronal classification quality. Journal of Neuroscience Methods. 2002;122:43–57. doi: 10.1016/s0165-0270(02)00276-5. [DOI] [PubMed] [Google Scholar]
- 56.Harris KD, Hirase H, Leinekugel X, Henze DA, Buzsaki G. Temporal interaction between single spikes and complex spike bursts in hippocampal pyramidal cells. Neuron. 2001;32:141–149. doi: 10.1016/s0896-6273(01)00447-0. [DOI] [PubMed] [Google Scholar]
- 57.Fee MS, Mitra PP, Kleinfeld D. Variability of extracellular spike waveforms of cortical neurons. J Neurophysiol. 1996;76:3823–3833. doi: 10.1152/jn.1996.76.6.3823. [DOI] [PubMed] [Google Scholar]
- 58.Quirk MC, Wilson MA. Interaction between spike waveform classification and temporal sequence detection. J.Neurosci.Methods. 1999;94:41–52. doi: 10.1016/s0165-0270(99)00124-7. [DOI] [PubMed] [Google Scholar]
- 59.Cohen M, Kohn A. Measuring and interpreting neuronal correlations. Nature Neuroscience. 2011;14:811–809. doi: 10.1038/nn.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pazienti A, Grun S. Robustness of the significance of spike synchrony withrespect to sorting errors. Journal of Computational Neuroscience. 2006;21:329–342. doi: 10.1007/s10827-006-8899-7. [DOI] [PubMed] [Google Scholar]
- 61.Ventura V, Gerkin RC. Accurately estimating neuronal correlation requiresa new spike-sorting paradigm. Proceedings of the National Academy of Sciences. 2012;109:7230–7235. doi: 10.1073/pnas.1115236109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganguly K, Dimitrov D, Wallis J, Carmena JM. Reversible large-scale modification of cortical networks during neuroprosthetic control. Nature Neuroscience. 2011;14:662–667. doi: 10.1038/nn.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fraser G, Chase S, Whitford A, Schwartz AB. Control of a brain-computer interface without spike sorting. Journal of Neural Engineering. 2009;6:055004. doi: 10.1088/1741-2560/6/5/055004. [DOI] [PubMed] [Google Scholar]
- 64.Redish AD, et al. Independence of firing corelates of anatomically proximate hippocampal pyramidal cells. The Journal of Neuroscience. 2001;21:1–6. doi: 10.1523/JNEUROSCI.21-05-j0004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shoham S, O'Connor DH, Segev R. How silent is the brain: is there a “dark matter” problem in neuroscience? J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2006;192:777–784. doi: 10.1007/s00359-006-0117-6. [DOI] [PubMed] [Google Scholar]
- 66.Moffitt MA, McIntyre CC. Model-based analysis of cortical recording with silicon microelectrodes. Clinical Neurophysiology. 2005;116:2240–2250. doi: 10.1016/j.clinph.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 67.Claverol-Tinture E, Nadasdy Z. Intersection of microwire electrodes with proximal CA1 stratum-pyramidale neurons at insertion for multi-unit recordings predictable by a 3D computer model. IEEE Trans Biomed Eng. 2004;51:2211–2216. doi: 10.1109/TBME.2004.834274. [DOI] [PubMed] [Google Scholar]
- 68.Buzsaki G, Mizuseki K. The log-dynamic brain: how skewed distributions affect network operations. Nat Rev Neurosci. 2014;15:264–278. doi: 10.1038/nrn3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Kock CP, Bruno RM, Spors H, Sakmann B. Layer- and cell-type-specific suprathreshold stimulus representation in rat primary somatosensory cortex. J Physiol. 2007;581:139–154. doi: 10.1113/jphysiol.2006.124321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakata S, Harris KD. Laminar structure of spontaneous and sensory-evoked population activity in auditory cortex. Neuron. 2009;64:404–418. doi: 10.1016/j.neuron.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niell CM, Stryker MP. Highly selective receptive fields in mouse visual cortex. J Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'Connor DH, Peron SP, Huber D, Svoboda K. Neural activity in barrel cortex underlying vibrissa-based object localization in mice. Neuron. 2010;67:1048–1061. doi: 10.1016/j.neuron.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 73.Hubel D. Tungsten microelectrode for recording from single units. Science. 1957;125:549–550. doi: 10.1126/science.125.3247.549. [DOI] [PubMed] [Google Scholar]
- 74.Kentros C, et al. Abolition of long-term stability of new hippocampal place cell maps by NMDA receptor blockade. Science. 1998;280:2121–2126. doi: 10.1126/science.280.5372.2121. [DOI] [PubMed] [Google Scholar]
- 75.Okun M, Carandini M, Harris KD. Long term recordings with immobile silicon probes in the mouse cortex. bioRxiv. 2015 doi: 10.1371/journal.pone.0151180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spruston N, Schiller Y, Stuart G, Sakmann B. Activity-dependent action potential invasion and calcium influx into hippocampal CA1 dendrites. Science. 1995;268:297–300. doi: 10.1126/science.7716524. [DOI] [PubMed] [Google Scholar]
- 77.Buzsaki G, Kandel A. Somadendritic backpropagation of action potentials in cortical pyramidal cells of the awake rat. J Neurophysiol. 1998;79:1587–1591. doi: 10.1152/jn.1998.79.3.1587. [DOI] [PubMed] [Google Scholar]
- 78.Tolias AS, et al. Recording chronically from the same neurons in awake, behaving primates. J Neurophysiol. 2007;98:3780–3790. doi: 10.1152/jn.00260.2007. [DOI] [PubMed] [Google Scholar]
- 79.Swindale NV, Spacek MA. Spike sorting for polytrodes: a divide and conquer approach. Front Syst Neurosci. 2014;8:6. doi: 10.3389/fnsys.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bar-Hillel A, Spiro A, Stark E. Spike sorting: Bayesian clustering of non-stationary data. Journal of Neuroscience Methods. 2006;157:303–316. doi: 10.1016/j.jneumeth.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 81.Jackson A, Fetz EE. Compact Movable Microwire Array for Long-Term Chronic Unit Recording in Cerebral Cortex of Primates. Journal of Neurophysiology. 2007;98:3109–3118. doi: 10.1152/jn.00569.2007. [DOI] [PubMed] [Google Scholar]
- 82.Fee MS, Mitra PP, Kleinfeld D. Automatic sorting of multiple unit neuronal signals in the presence of anisotropic and non-Gaussian variability. J Neurosci Methods. 1996;69:175–188. doi: 10.1016/S0165-0270(96)00050-7. [DOI] [PubMed] [Google Scholar]
- 83.Williams JC, Rennaker RL, Kipke DR. Stability of chronic multichannel neural recordings: Implications for a long-term neural interface. Neurocomputing. 1999;26–7:1069–1076. [Google Scholar]
- 84.Greenberg PA, Wilson FA. Functional stability of dorsolateral prefrontal neurons. Journal of Neurophysiology. 2004;92:1042–1055. doi: 10.1152/jn.00062.2004. [DOI] [PubMed] [Google Scholar]
- 85.Ohki K, Chung S, Ch'ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- 86.Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Theis L, et al. Benchmarking Spike Rate Inference in Population Calcium Imaging. Neuron. 2016;90:471–482. doi: 10.1016/j.neuron.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rose T, Goltstein PM, Portugues R, Griesbeck O. Putting a finishing touch on GECIs. Frontiers in molecular neuroscience. 2014;7:88. doi: 10.3389/fnmol.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghosh KK, et al. Miniaturized integration of a fluorescence microscope. Nature methods. 2011;8:871–878. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Keller PJ, Ahrens MB, Freeman J. Light-sheet imaging for systems neuroscience. Nature methods. 2015;12:27–29. doi: 10.1038/nmeth.3214. [DOI] [PubMed] [Google Scholar]
- 91.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ohki K, et al. Highly ordered arrangement of single neurons in orientation pinwheels. Nature. 2006;442:925–928. doi: 10.1038/nature05019. [DOI] [PubMed] [Google Scholar]
- 93.Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yasuda R, et al. Imaging calcium concentration dynamics in small neuronal compartments. Sci STKE. 2004;2004:pl5. doi: 10.1126/stke.2192004pl5. [DOI] [PubMed] [Google Scholar]
- 95.Podgorski K, Ranganathan G. Brain heating induced by near infrared lasers during multiphoton microscopy. bioRxiv. 2016 doi: 10.1152/jn.00275.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Valmianski I, et al. Automatic identification of fluorescently labeled brain cells for rapid functional imaging. J Neurophysiol. 2010;104:1803–1811. doi: 10.1152/jn.00484.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sadovsky AJ, et al. Heuristically optimal path scanning for high-speed multiphoton circuit imaging. J Neurophysiol. 2011;106:1591–1598. doi: 10.1152/jn.00334.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cotton RJ, Froudarakis E, Storer P, Saggau P, Tolias AS. Three-dimensional mapping of microcircuit correlation structure. Frontiers in neural circuits. 2013;7:151. doi: 10.3389/fncir.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hamel EJ, Grewe BF, Parker JG, Schnitzer MJ. Cellular level brain imaging in behaving mammals: an engineering approach. Neuron. 2015;86:140–159. doi: 10.1016/j.neuron.2015.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wilt BA, Fitzgerald JE, Schnitzer MJ. Photon shot noise limits on optical detection of neuronal spikes and estimation of spike timing. Biophys J. 2013;104:51–62. doi: 10.1016/j.bpj.2012.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Young MD, Field JJ, Sheetz KE, Bartels RA, Squier J. A pragmatic guide to multiphoton microscope design. Adv Opt Photonics. 2015;7:276–378. doi: 10.1364/AOP.7.000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kerr JN, Greenberg D, Helmchen F. Imaging input and output of neocortical networks in vivo. Proc.Natl.Acad.Sci.U.S.A. 2005;102:14063–14068. doi: 10.1073/pnas.0506029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lecoq J, et al. Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging. Nat Neurosci. 2014;17:1825–1829. doi: 10.1038/nn.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stirman JN, Smith IT, Kudenov MW, Smith SL. Wide field-of-view, twin-region two-photon imaging across extended cortical networks. biorxiv.org. 2014 [Google Scholar]
- 105.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nature methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 106.Yang W, et al. Simultaneous Multi-plane Imaging of Neural Circuits. Neuron. 2016;89:269–284. doi: 10.1016/j.neuron.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dunn AK, Wallace VP, Coleno M, Berns MW, Tromberg BJ. Influence of optical properties on two-photon fluorescence imaging in turbid samples. Appl Opt. 2000;39:1194–1201. doi: 10.1364/ao.39.001194. [DOI] [PubMed] [Google Scholar]
- 108.Tung CK, et al. Effects of objective numerical apertures on achievable imaging depths in multiphoton microscopy. Microsc Res Tech. 2004;65:308–314. doi: 10.1002/jemt.20116. [DOI] [PubMed] [Google Scholar]
- 109.Zheng G, Ou X, Horstmeyer R, Yang C. Characterization of spatially varying aberrations for wide field-of-view microscopy. Opt Express. 2013;21:15131–15143. doi: 10.1364/OE.21.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.von Diezmann A, Lee MY, Lew MD, Moerner WE. Correcting field-dependent aberrations with nanoscale accuracy in three-dimensional single-molecule localization microscopy. Optica. 2015;2:985–993. doi: 10.1364/OPTICA.2.000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang C, Ji N. Characterization and improvement of three-dimensional imaging performance of GRIN-lens-based two-photon fluorescence endomicroscopes with adaptive optics. Opt Express. 2013;21:27142–27154. doi: 10.1364/OE.21.027142. [DOI] [PubMed] [Google Scholar]
- 112.Barretto RP, Messerschmidt B, Schnitzer MJ. In vivo fluorescence imaging with high-resolution microlenses. Nature methods. 2009;6:511–512. doi: 10.1038/nmeth.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ji N, Sato TR, Betzig E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc Natl Acad Sci U S A. 2012;109:22–27. doi: 10.1073/pnas.1109202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dana H, et al. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS One. 2014;9:e108697. doi: 10.1371/journal.pone.0108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen Q, et al. Imaging neural activity using Thy1-GCaMP transgenic mice. Neuron. 2012;76:297–308. doi: 10.1016/j.neuron.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Glickfeld LL, Andermann ML, Bonin V, Reid RC. Cortico-cortical projections in mouse visual cortex are functionally target specific. Nat Neurosci. 2013;16:219–226. doi: 10.1038/nn.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peron SP, Freeman J, Iyer V, Guo C, Svoboda K. A Cellular Resolution Map of Barrel Cortex Activity during Tactile Behavior. Neuron. 2015;86:783–799. doi: 10.1016/j.neuron.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 118.Bonin V, Histed MH, Yurgenson S, Reid RC. Local diversity and fine-scale organization of receptive fields in mouse visual cortex. J Neurosci. 2011;31:18506–18521. doi: 10.1523/JNEUROSCI.2974-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen JL, Pfaffli OA, Voigt FF, Margolis DJ, Helmchen F. Online correction of licking-induced brain motion during two-photon imaging with a tunable lens. J Physiol. 2013;591:4689–4698. doi: 10.1113/jphysiol.2013.259804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Holtmaat A, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dubbs A, Guevara J, Yuste R. moco: Fast Motion Correction for Calcium Imaging. Frontiers in neuroinformatics. 2016;10:6. doi: 10.3389/fninf.2016.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 124.Greenberg DS, Kerr JN. Automated correction of fast motion artifacts for two-photon imaging of awake animals. J Neurosci Methods. 2009;176:1–15. doi: 10.1016/j.jneumeth.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 125.Wilms CD, Hausser M. Reading out a spatiotemporal population code by imaging neighbouring parallel fibre axons in vivo. Nat Commun. 2015;6:6464. doi: 10.1038/ncomms7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dufour P, Piche M, De Koninck Y, McCarthy N. Two-photon excitation fluorescence microscopy with a high depth of field using an axicon. Appl Opt. 2006;45:9246–9252. doi: 10.1364/ao.45.009246. [DOI] [PubMed] [Google Scholar]
- 127.Andermann ML, Kerlin AM, Reid RC. Chronic cellular imaging of mouse visual cortex during operant behavior and passive viewing. Frontiers in cellular neuroscience. 2010;4:3. doi: 10.3389/fncel.2010.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Laffray S, et al. Adaptive movement compensation for in vivo imaging of fast cellular dynamics within a moving tissue. PLoS One. 2011;6:e19928. doi: 10.1371/journal.pone.0019928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Andermann ML, Kerlin AM, Roumis DK, Glickfeld LL, Reid RC. Functional specialization of mouse higher visual cortical areas. Neuron. 2011;72:1025–1039. doi: 10.1016/j.neuron.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schultz SR, Kitamura K, Post-Uiterweer A, Krupic J, Hausser M. Spatial pattern coding of sensory information by climbing fiber-evoked calcium signals in networks of neighboring cerebellar Purkinje cells. J Neurosci. 2009;29:8005–8015. doi: 10.1523/JNEUROSCI.4919-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pnevmatikakis EA, et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron. 2016;89:285–299. doi: 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Andilla FD, Hamprecht FA. Advances in Neural Information Processing Systems. 2014. pp. 64–72. [Google Scholar]
- 135.Pachitariu M, et al. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. BioRxiv. 2016;061507 [Google Scholar]
- 136.Pachitariu M, et al. Advances in Neural Information Processing Systems. 2013. pp. 1745–1753. [Google Scholar]
- 137.Vogelstein JT, et al. Fast nonnegative deconvolution for spike train inference from population calcium imaging. J Neurophysiol. 2010;104:3691–3704. doi: 10.1152/jn.01073.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yaksi E, Friedrich RW. Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging. Nature methods. 2006;3:377–383. doi: 10.1038/nmeth874. [DOI] [PubMed] [Google Scholar]
- 139.Ganmore E, Krumin M, Rossi LF, Carandini M, Simoncelli EP. Direct estimation of firing rates from calcium imaging data. arxiv.org. 2016 [Google Scholar]
- 140.Harvey CD, Coen P, Tank DW. Choice-specific sequences in parietal cortex during a virtual-navigation decision task. Nature. 2012;484:62–68. doi: 10.1038/nature10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Smith SL, Hausser M. Parallel processing of visual space by neighboring neurons in mouse visual cortex. Nat Neurosci. 2010;13:1144–1149. doi: 10.1038/nn.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Harris KD, Csicsvari J, Hirase H, Dragoi G, Buzsaki G. Organization of cell assemblies in the hippocampus. Nature. 2003;424:552–556. doi: 10.1038/nature01834. [DOI] [PubMed] [Google Scholar]