Abstract
Growth factor receptor bound protein 7 (Grb7) is a signal transducing adaptor protein that mediates specific protein-protein interactions in multiple signaling pathways. Grb7, with Grb10 and Grb14, are members of the Grb7 protein family. The topology of the Grb7 family members contains several protein-binding domains that facilitate the formation of protein complexes and high signal transduction efficiency. Grb7 has been found overexpressed in several types of cancers and cancer cell lines, and is presumed involved in cancer progression through promotion of cell proliferation and migration via interactions with the ErbB2 (HER2) receptor, FAK (focal adhesion kinase), Ras-GTPases, and other signaling partners. We previously reported Grb7 binds to Hax1 (HS1 associated protein X1) isoform 1, an anti-apoptotic protein also involved in cell proliferation and calcium homeostasis. In this study, we confirm the in vitro Grb7/Hax1 interaction is exclusive to these two proteins and their interaction does not depend on Grb7 dimerization state. In addition, we report Grb7 and Hax1 isoform 1 may colocalize partially to mitochondria in EGF treated SKBR3 cells and growth conditions can affect this colocalization. Moreover, Grb7 can affect Caspase3 cleavage of the Hax1 isoform 1 in vitro, and Grb7 expression may slow the Caspase3 cleavage of Hax1 isoform 1 in apoptotic HeLa cells. Finally, Grb7 is shown to increase cell viability in apoptotic HeLa cells in a time dependent manner. Taken together, these discoveries provide clues for the role of a Grb7/Hax1 protein interaction in apoptosis pathways involving Hax1.
Keywords: signal transducing adaptor protein, Grb7, Hax1, mitochondria, SKBR3 cells, apoptosis, Caspase3
Introduction
Growth factor receptor bound protein 7 (Grb7) is a member of the Grb7 signaling adaptor protein family, whose members are Grb7, Grb10 and Grb14 (Daly et al. 1996; Frantz et al. 1997; Margolis et al. 1992; Ooi et al. 1995). The Grb7 protein family shares a highly conserved domain topology composed of an N-terminal Proline rich region, an RA (Ras Associating) domain, a central PH (Pleckstrin Homology) domain, a BPS (Between PH and SH2 domains) motif, and a C-terminal SH2 (Src Homology 2) domain (Daly 1998; Han et al. 2001; Holt and Siddle 2005; Morrione 2000; Shen and Guan 2004) (Figure 1-A top). The RA-PH domains and BPS motif together are also known as the GM region (Grb and Mig homology region) because this region shares homology with the corresponding region in the C. elegans neuronal cell migration protein Mig10 (Manser et al. 1997; Ooi et al. 1995; Stein et al. 1994).
Figure 1. A) Top: Domain topology of the human Grb7 protein isoform 1.
The approximate amino acid residue numbers defining each domain are indicated by numbers. Bottom: Domain topology of the human Hax1 protein isoform 1 The approximate amino acid residue numbers defining each domain or motif are indicated by numbers. B) Western Blot results for the in vitro binding assay of purified SUMO-Grb7-RAPH domains and purified Hax1 Lane 1: Last wash sample from the negative control. Lane 2: Last wash sample from the in vitro binding assay of SUMO-RAPH and Hax1. Lane 3: Blank. Lane 4–6: Elution samples (three elutions) from the in vitro binding assay of SUMO-RAPH and Hax1. Lane 7–9: Elution samples (three elutions) from the negative control. C) Western Blot results for the in vitro binding assay of purified Grb7 or Grb7 (F511R) and purified Hax1 Lane 1: Last wash sample from the in vitro binding assay of Grb7 and Hax1. Lane 2: Last wash sample from the in vitro binding assay of Grb7 (F511R) and Hax1. Lane 3: Last wash sample from the negative control. Lane 4–5: Elution samples (two elutions) from the in vitro binding assay of Grb7 and Hax1. Lane 6: Blank. Lane 7–8: Elution samples (two elutions) from the in vitro binding assay of Grb7 (F511R) and Hax1. Lane 9: Blank. Lane 10–11: Elution samples (two elutions) from the negative control.
The multiple domain structure of the Grb7 protein permits it to take part in a variety of signal transduction pathways (Han et al. 2001; Holt and Siddle 2005; Shen and Guan 2004). Grb7 binds to the ErbB receptor family, PDGF (platelet-derived growth factor) receptor, FAK (focal adhesion kinase) and insulin receptor through its SH2 domain (Chu et al. 2009; Fiddes et al. 1998; Han and Guan 1999; Kasus-Jacobi et al. 2000; Margolis et al. 1992; Stein et al. 1994; Yokote et al. 1996). To some degree Grb7 binds to Ras-GTPases through its RA domain (Chu et al. 2010). Finally, Grb7 binds to PIP3 (Phosphatidyl Inositol-3-Phosphate) phospholipid through its PH domain (Shen et al. 2002). Our own laboratory has reported Grb7 interactions with FHL2 (Four and a Half LIM domains isoform 2), Filamin-α and Hax1 through its central Grb7-RAPH domain region (Paudyal et al. 2013; Siamakpour-Reihani et al. 2011; Siamakpour-Reihani et al. 2009).
Hax1 (HS1 associated protein X1) was originally shown to interact with HS1, a Src kinase substrate (Suzuki et al. 1997). Hax1 is a multifunctional protein involved in cell proliferation, calcium homeostasis, and regulation of apoptosis; an often deregulated process in carcinogenesis (Cavnar et al. 2011; Cilenti et al. 2004; Han et al. 2006; Kang et al. 2010; Lee et al. 2008; Radhika et al. 2004; Ramsay et al. 2007; Vafiadaki et al. 2007; Vafiadaki et al. 2009; Yap et al. 2010). The protein displays two disputed Bcl-2 Homology domains termed BH1 and BH2, a PEST motif for targeting of the protein for proteasomic degradation, and a disputed C-terminal transmembrane domain (Chao et al. 2008; Jeyaraju et al. 2009; Li et al. 2012; Sharp et al. 2002; Suzuki et al. 1997) (Figure 1-A bottom). The human Hax1 protein has different isoforms and Hax1 isoform 1 is the major observed form (Grzybowska et al. 2013). The rat Hax1 isoform 1 (the human Hax1 isoform 1 homologue) forms a strong homodimer with a dissociation constant (Kd) of approximately 3.8nM (Koontz and Kontrogianni-Konstantopoulos 2014).
A series of experiments have confirmed the anti-apoptotic effects and cell-protective properties of Hax1 isoform 1 (Chao et al. 2008; Han et al. 2006; Sharp et al. 2002). As well, overexpression of Hax1 has been observed in several cancers, including breast, lung, and melanoma (Trebinska et al. 2010). Although Hax1 does not have a recognized protein interaction domain structure like Grb7, it has large numbers of binding partners, of which some are active in apoptosis signaling (Cilenti et al. 2004; Han et al. 2006; Kang et al. 2010; Lee et al. 2008; Matsuda et al. 2003; Ruzzene et al. 2002; Vafiadaki et al. 2007; Vafiadaki et al. 2009). Hax1 can bind to Caspase9 and inhibit Caspase9 activities (Han et al. 2006), and can also be cleaved by Caspase3 upon apoptosis (Lee et al. 2008). Hax1 binds to XIAP (X-linked inhibitor of apoptosis) and prevents XIAP from polyubiquitination (Kang et al. 2010). Hax1 also interacts with the calcium pump sarco(endo)plasmic reticulum calcium transport ATPase2 (SERCA2) and its inhibitor phospholamban (PLN). In these roles Hax1 can mediate calcium homeostasis and apoptosis regulation (Vafiadaki et al. 2007; Vafiadaki et al. 2009).
In contrast to Hax1, previous studies linking Grb7 to apoptosis signaling have been sparse (Giricz et al. 2012; Han et al. 2001). Grb7 depletion or inhibition in TNBC (Triple-Negative Breast Cancer) cell lines may promote cell death by apoptosis in 3D extracellular matrix cultures, but the mechanism is still not fully understood (Giricz et al. 2012). We previously reported Grb7 interacts with Hax1, which suggests the interaction could alter each protein’s signaling function. The current studies herein seek to confirm direct and exclusive binding between Grb7 and Hax1, and provide a first exploration of the possible role of this interaction in apoptosis pathways.
Materials and Methods
Materials
The anti-SUMO (mouse) monoclonal antibody (product number 200-301-428) was purchased from Rockland Immunochemicals (Limerick, PA). The anti-Hax1 (mouse) monoclonal antibody (product number 610825) and Ac-DEVD-AMC (product number 556449) were purchased from BD Biosciences (San Jose, CA). The anti-Grb7 (rabbit) polyclonal antibody (H-70) (product number sc-13954), anti-Caspase3 p17 (mouse) monoclonal antibody (B-4) (product number sc-271028), anti-Actin (mouse) monoclonal antibody (C-2) (product number sc-8432), goat anti-mouse IgG-HRP (product number sc-2005) and goat anti-rabbit IgG-HRP (product number sc-2004) were purchased from Santa Cruz Biotechnology (Dallas, TX). The Pierce Co-Immunoprecipitation Kit (product number 26149) and Pierce Antibody Clean-up Kit (product number 44600) were obtained from Thermo Fisher Scientific (Waltham, MA). Lipofectamine 3000 (product number L3000-001), Lipofectamine LTX Reagent with PLUS Reagent (product number A12621), MitoTracker Deep Red FM dye (product number M22426), Lectin GS-II Alexa Fluor 647 Conjugate (product number L32451), F(ab’)2-Goat anti-Rabbit IgG (H+L) Secondary Antibody Alexa Fluor 488 conjugate (product number A11070), Goat anti-Mouse IgG (H+L) Secondary Antibody Alexa Fluor 546 conjugate (product number A11030) and Hoechst 33342 (product number H3570) were obtained from Life Technologies (Grand Island, NY). CellTiter-Glo Luminescent Cell Viability Assay (product number G7570) was from Promega Corporation (Madison, WI). Staurosporine (product number ab120056) was from Abcam (Cambridge, MA). Etoposide (product number E1383) was from Sigma-Aldrich (St Louis, MO).
Methods
DNA Constructs and Plasmids
The pDNR-LIB-Hax1 plasmid (Siamakpour-Reihani et al. 2011) was used as a template for amplification of the full length human Hax1 gene (NCBI Reference Sequence: NM_006118.3 transcript variant 1; UniProt: O00165 isoform 1) by PCR (Forward primer: 5’ ct gga tcc ATG AGC CTC TTT GAT CTC TTC C, Reverse primer: 5’ tat gaa ttc CTA CCG GGA CCG GAA CCA AC). The resulting PCR fragment was digested by the endonucleases BamHI and EcoRI, followed by ligation into the pGEX-2T vector to generate the protein expression plasmid pGEX-2T-Hax1.
Using the pGEX-2T-Hax1 plasmid as a template, the coding sequence for full length human Hax1 was amplified by PCR (Forward primer: 5’ ga cag tcg cat ATG AGC CTC TTT GAT CTC TTC CG, Reverse primer: 5’gt ata tct aag ctt TAA CCG GGA CCG GAA CCA ACG). The PCR fragment was digested by the endonucleases NdeI and Hind III (HF), followed by ligation into the pET22b(+) vector (product number 69744-3) from EMD Millipore (Billerica, MA) to generate the protein expression plasmid pET22b(+)-Hax1.
The pET303-RAPH plasmid (Siamakpour-Reihani et al. 2009) was used as a template for amplification of the human Grb7-RAPH domains (NCBI Reference Sequence: NM_005310.3 transcript variant 1; Uniprot: Q14451 isoform 1 residues 100 to 338) by PCR (Forward primer: 5’ at cgt ctc t a ggt CGC CCC CAT GTA GTA AAG, Reverse primer: 5’ cg tct aga tca GTA CTT GAA GAG GCG GAA GG). The resulting PCR fragment was digested by the endonucleases BsmBI and XbaI, followed by ligation into the pE-SUMOpro vector (product number 1001A) from LifeSensors Inc. (Malvern, PA) to generate the protein expression plasmid pE-SUMOpro-RAPH.
The pCMV-Myc-Grb7 plasmid (Siamakpour-Reihani et al. 2009) was used as a template for amplification of the full length human Grb7 gene by PCR (Forward primer: 5’ g aga tct aga ATG GAG CTG GAT CTG TCT CCA CCT CAT CTT, Reverse primer: 5’ g aga ctc gag GAG GGC CAC CCG CGT GC). The resulting PCR fragment was digested by the endonucleases XbaI and XhoI, followed by ligation into the pET303 vector to produce the protein expression plasmid pET303-Grb7.
The pET303-Grb7 plasmid was used as a template to produce the single-site mutant pET303-Grb7 (F511R) expression plasmid using the QuikChange II site-directed mutagenesis kit (product number 200523) from Agilent Technologies (Santa Clara, CA) according to the manufacturer’s protocol. Primers were designed using the online tool at http://www.genomics.agilent.com/primerDesignProgram.jsp.
The plasmid pcDNA3.1(+)/Luc2=tdT was received as a gift from the Christopher Contag Lab (Addgene plasmid product number 32904) (Patel et al. 2010). The plasmid was digested using the endonuclease EcoRI (HF) to remove the luc2=tdT (Firefly Luciferase-tandem Tomato Red Fluorescent Protein) gene insert. The digested pcDNA3.1(+) vector was self-ligated by T4 DNA Ligase to generate an intact pcDNA3.1(+) plasmid.
The pET22b(+)-Hax1 plasmid was used as a template for amplification of the full length human Hax1 gene by PCR (Forward primer: 5’ tca gga tcc ACC ATG AGC CTC TTT GAT CTC TTC CG; Reverse primer: 5’ ata cat ctc gaa ttc CTA CCG GGA CCG GAA CCA AC). The resulting PCR fragment was digested by endonucleases BamHI (HF) and EcoRI (HF), followed by ligation into pcDNA3.1(+) to generate the plasmid pcDNA3.1(+)-Hax1.
The pET303-Grb7 plasmid was used as a template for amplification of the full length human Grb7 gene by PCR (Forward primer: 5’ ctc gaa ttc ACC ATG GAG CTG GAT CTG TCT CCA, Reverse primer: 5’ gta tca cta tct aga TTA GAG GGC CAC CCG CGT). The resulting PCR fragment was digested by endonucleases EcoRI (HF) and XbaI followed by ligation into pcDNA3.1(+) to generate the plasmid pcDNA3.1(+)-Grb7.
The pET23b(+)-ProCaspase3 plasmid was received as a gift from the Guy Salvesen Lab (Addgene plasmid product number 11821) (Zhou et al. 1997). The plasmid was used as a template for amplification of the full length human ProCaspase3 gene (NCBI Reference Sequence: NM_004346.3 transcript variant alpha; Uniprot P42574) by PCR (Forward primer: 5’ a tat ata gga tcc ACC ATG GAG AAC ACT GAA, Reverse primer: 5’ ctg cac gaa ttc TTA GTG ATA AAA ATA GAG TTC T). The resulting PCR fragment was digested by endonucleases BamHI (HF) and EcoRI (HF) followed by ligation into pcDNA3.1(+) to generate the plasmid pcDNA3.1(+)-ProCaspase3.
Protein Expression and Purification
Protein Expression
For all protein expression plasmids were transformed into E. coli Rosetta 2(DE3) competent cells (product number 71400-4) from EMD Millipore (Billerica, MA).
The expression of the Hax1-6xHis, Grb7-RAPH-6xHis, 6xHis-SUMO, 6xHis-SUMO-Grb7-RAPH, full-length-Grb7-6xHis (FL-Grb7-6xHis), FL-Grb7 (F511R)-6xHis, GST and GST-Hax1 proteins follow a general similar procedure. Transformed bacterial cells were grown in LB medium with appropriate antibiotics (100µg/mL Ampicillin, 35µg/mL Chloramphenicol) at 37°C with shaking at 225rpm until cell growth reached an approximate optical density (OD600) of 0.6. Protein expression was then induced with 0.5mM IPTG at 16°C and shaking at 200rpm overnight. Bacterial cells were harvested by centrifugation at 9715 × g and 4°C for 15 minutes, and the resulting cell pellets were stored at −20°C until further use.
The expression of Caspase3-6xHis was described previously (Denault and Salvesen 2003; Zorn et al. 2012). Transformed bacterial cells were grown in 2xYT medium (100µg/mL Ampicillin, 35µg/mL Chloramphenicol) at 37°C with shaking at 225rpm until an OD600 of 0.7 was achieved. Protein expression was then induced with 0.2mM IPTG at 30°C and 200rpm shaking for 6 hours. Bacterial cells were harvested by centrifugation at 9715 × g and 4°C for 15 minutes, and the resulting cell pellets were stored at −20°C until further use.
Protein Purification
The purification of the Hax1-6xHis, Grb7-RAPH-6xHis, 6xHis-SUMO, 6xHis-SUMO-Grb7-RAPH, FL-Grb7-6xHis, FL-Grb7 (F511R)-6xHis and Caspase3-6xHis proteins follow a general similar procedure. Bacterial cell pellets were resuspended in lysis buffer (25mM Tris-HCl, 500mM NaCl, 5% w/v Glycerol, 20mM Imidazole, 0.5µg/mL Leupeptin, 0.5µg/mL Pepstatin A, 0.05mM PMSF, 20mM β-ME, pH8.0), and sonicated on ice for 5 × 12 seconds (60% amplitude). Tween-20 was added to the sonicated cell lysate to 1% w/v, and the mixture was gently rocked at 4°C for 30 minutes. After centrifugation at 5465 × g at 4°C for 15 minutes, the cleared cell lysate was collected and combined with Ni-NTA resins. The lysate/Ni-NTA mixture was gently rocked at 4°C for 2 hours, followed by transfer to a gravity flow column. The settled Ni-NTA resins were washed with wash buffer (25mM Tris-HCl, 600mM NaCl, 5% w/v Glycerol, 20mM to 100mM Imidazole, 1% w/v Tween-20, pH 8.0) for multiple times. Proteins were eluted from the Ni-NTA resin with elution buffer (25mM Tris-HCl, 300mM NaCl, 5% w/v Glycerol, 200mM Imidazole, 1% w/v Tween-20, pH 8.0).
For the Grb7-6xHis and Grb7 (F511R)-6xHis proteins, the Ni-NTA resin elutions were precipitated by PEG 6000 and Ammonium Sulfate for further purification. Specifically, 50% w/v PEG 6000 aqueous solution was added to the eluted protein solution to 5% w/v. The mixture was then gently rocked or stirred at 4°C for 30 minutes. After centrifugation at 9000 × g at 4°C for 15 minutes, the pellet of protein was immediately resuspended in resuspension buffer (25mM Tris-HCl, 300mM NaCl, 5% w/v Glycerol, 0.1% w/v Tween-20, pH 8.0). To promote solvation, the resuspended protein solution was gently rocked or stirred at 4°C overnight. After centrifugation at 9000 × g at 4°C for 15 minutes, the supernatant with dissolved protein was collected for the following Ammonium Sulfate precipitation: 3M Ammonium Sulfate/resuspension buffer solution was immediately added to the solution with dissolved protein to 1M. The mixture was then gently rocked or stirred at 4°C for 30 minutes. After centrifugation at 9000 × g at 4°C for 15 minutes, the pellet of protein was immediately resuspended and readily dissolved in the resuspension buffer. For following assays, the purified Grb7-6xHis and Grb7 (F511R)-6xHis proteins were dialyzed into 20mM HEPES, 100mM NaCl, 5% w/v Glycerol, 0.1% w/v Tween-20, pH 7.5.
For further purification, the Hax1-6xHis protein Ni-NTA resin elutions were precipitated using Ammonium Sulfate. Ammonium Sulfate precipitation procedures were as described previously, with the Ammonium Sulfate solution concentration modified to 0.4M. For the following assays, purified Hax1-6xHis protein was dialyzed into 20mM HEPES, 100mM NaCl, 5% w/v Glycerol, 0.1% w/v Tween-20, pH 7.5.
For the Grb7-RAPH-6xHis protein purification, Ni-NTA resin elutions were immediately dialyzed into 20mM HEPES, 100mM NaCl, 5% w/v Glycerol, 1% w/v Tween-20, pH 7.5.
The purifications of the GST and GST-Hax1 proteins followed a similar procedure. Bacterial cell pellets were resuspended in lysis/wash buffer (1xPBS, 0.5µg/mL Leupeptin, 0.5µg/mL Pepstatin A, 0.05mM PMSF, 10mM DTT, pH 7.4), and then sonicated on ice for 5 × 12 seconds. Tween-20 was added to the cell lysate to 1% w/v, and the mixture was gently rocked at 4°C for 30 minutes. After centrifugation at 5465 × g at 4°C for 15 minutes, the cleared cell lysate was collected and combined with Glutathione Sepharose 4B (GluSeph) agarose beads, followed by gentle rocking at 4°C for 2 hours. The above mixture was transferred to a gravity flow column and the settled GluSeph beads were washed with lysis/wash buffer (1xPBS, 0.5µg/mL Leupeptin, 0.5µg/mL Pepstatin A, 0.05mM PMSF, 10mM DTT, pH 7.4) for multiple times. Proteins were eluted from the GluSeph beads with elution buffer (50mM Tris-HCl, 20mM reduced Glutathione, pH 8.0).
Antibody Immobilization
The anti-SUMO (mouse) monoclonal antibody in 1xPBS and anti-Grb7 (rabbit) polyclonal antibody (H-70) in 1xPBS were used for immobilization. Prior to immobilization, a Pierce Antibody Clean-up Kit was used to remove gelatin from the anti-Grb7 (rabbit) polyclonal antibody (H-70). The antibodies were covalently immobilized to AminoLink Plus Coupling Resins using the Pierce Co-Immunoprecipitation method according to the manufacturer’s protocol (Thermo Fisher Scientific).
in vitro Binding Assay
Equimolar amounts of 6xHis-SUMO (2.48µg, 0.5µM) and 6xHis-SUMO-Grb7-RAPH (7.92µg, 0.5µM) proteins were separately incubated with Hax1-6xHis (3.30µg, 0.25µM) and anti-SUMO (mouse) monoclonal antibodies (20µg) bound to AminoLink Plus Coupling Resins (25µL bed volume) in IP lysis/wash buffer (25mM Tris-HCl, 150mM NaCl, 1mM EDTA, 1% w/v NP-40, 5% w/v glycerol, pH 7.4) (plus 1mM DTT, 0.02% w/v NaN3) (total volume: 400µL) with gentle rocking at 4°C overnight. After overnight incubation, the resins were washed and the protein complexes were eluted from resins following the Pierce Co-Immunoprecipitation method according to the manufacturer’s protocol (Thermo Fisher Scientific). The washed and eluted samples were analyzed by SDS-PAGE followed by Western Blot.
Equimolar amounts of Grb7-6xHis (10.90 µg, 0.45µM) and Grb7 (F511R)-6xHis (10.90 µg, 0.45µM) proteins were separately incubated with Hax1-6xHis (3.30µg, 0.25µM) and anti-Grb7 (rabbit) polyclonal antibody (15µg) bound to AminoLink Plus Coupling Resin (25µL bed volume) in IP lysis/wash buffer (25mM Tris-HCl, 150mM NaCl, 1mM EDTA, 1% w/v NP-40, 5% w/v glycerol, pH 7.4) (plus 1mM DTT, 0.02% w/v NaN3) (total volume 400µL) with gentle rocking at 4°C overnight. After overnight incubation, the resins were washed and the protein complexes were eluted from resins. The washed and eluted samples were analyzed by SDS-PAGE followed by Western Blot.
Cell Culture and Transfections
SKBR3 and HeLa cells (gifts from the Dr. Aaron Rowland Lab) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% v/v Fetal Bovine Serum (FBS) and grown in an incubator at 37°C with 5% CO2. Transient transfections were performed using Lipofectamine LTX or Lipofectamine 3000 transfection agent according to the manufacturer’s protocol (Life Technologies).
EGF stimulation studies were performed as follows. Cells were serum starved overnight, incubated with 10ng/mL EGF at 37°C and 5% CO2 for 15 to 30 minutes, followed by cell fixation and permeabilization for immunofluorescence analyses.
To prepare mammalian cell lysates for Western Blot, cells were washed first with 1xPBS, and then lysed with ice-cold IP lysis/wash buffer (25mM Tris-HCl, 150mM NaCl, 1mM EDTA, 1% w/v NP-40, 5% w/v glycerol, 1µg/mL Leupeptin, 1µg/mL Pepstatin A, 0.1mM PMSF, pH 7.4).
Immunofluorescence Analysis
SKBR3 cells were grown on glass bottom dishes from MatTek Corporation (Ashland, MA) and transfected with plasmid pcDNA3.1(+)-Hax1. Two days post-transfection cells were fixed with 3.7% v/v formaldehyde/1xPBS (15 minutes), permeabilized with 0.1% v/v Triton X-100/1xPBS (15 minutes), and blocked with 3% w/v BSA/0.05% w/v Tween-20/1xPBS (1 hour). Cells were then incubated with the diluted primary antibodies in 1% w/v BSA/0.05% w/v Tween-20/1xPBS (1:600-1:1000 dilution for anti-Grb7 (rabbit) polyclonal antibody (H-70), 1:1000 dilution for anti-Hax1 (mouse) monoclonal antibody) at 4°C overnight. Next, cells were washed with 1xPBS and incubated with diluted secondary antibodies and Hoechst 33342 (2µg/mL) in 1% w/v BSA/0.05% w/v Tween-20/1xPBS (1:400-1:500 dilution for F(ab’)2-Goat anti-Rabbit IgG (H+L) secondary antibody Alexa Fluor 488 conjugate, 1:500 for Goat anti-Mouse IgG (H+L) secondary antibody Alexa Fluor 546 conjugate) (1 hour). Finally, cells were washed with 1xPBS, immersed in glycerol, and stored at 4°C until further use. Images were acquired using a 63×1.4-0.6 oil objective lens equipped Leica TCS SP5 Confocal Laser Scanning Microscope.
Cellular mitochondria were visualized using the following protocol. Transfected cells were incubated with 200nM MitoTracker Deep Red FM dye at 37°C and 5% CO2 for 15 minutes. After staining, cells were washed with 1xPBS and fixed and permeabilized as described above.
Cellular golgi bodies were stained according to the subsequent protocol. Post-incubation with primary antibodies, cells were incubated with 5µg/mL Lectin GS-IIAlexa Fluor 647 Conjugate together with diluted secondary antibodies and Hoechst 33342 (1 hour) as described above.
in vitro Caspase3 Cleavage of Hax1 Assay
Equimolar amounts of Grb7-6xHis (6.08µg, 1µM), Grb7-RAPH-6xHis (2.84µg, 1µM) proteins and GST (2.70µg, 1µM) protein were separately incubated with Hax1-6xHis (0.33µg, 0.1µM) and Caspase3-6xHis (0.57µg, 0.1µM) in Caspase assay buffer (50mM HEPES, 100mM NaCl, 0.1% w/v CHAPS, 10mM DTT, 1mM EDTA, 10% w/v Glycerol, pH 7.4; final volume: 100µL) at 37°C for 1.5 hours. For each reaction, 15µL samples were collected every 30 minutes and analyzed by SDS-PAGE followed by Western Blot.
Caspase3 Activity Assay
Equimolar amounts of Grb7-6xHis (0.30 µg, 0.05µM), Grb7-RAPH-6xHis (0.14 µg, 0.05µM) and Hax1-6xHis (0.16 µg, 0.05µM) proteins were separately incubated with Caspase3-6xHis (0.03µg, 0.005µM) and Ac-DEVD-AMC (0.50µg, 7.4µM) in Caspase assay buffer (50mM HEPES, 100mM NaCl, 0.1% w/v CHAPS, 10mM DTT, 1mM EDTA, 10% w/v Glycerol, pH 7.4; total volume: 100µL) in one well of 96-well plate. Each reaction was repeated in triplicate using 3 wells of a 96-well plate. The plate was then immediately placed into a Bio-Tek FL600 fluorescence/absorbance plate reader and the fluorescence (Ex360nm/Em460nm) from liberated AMC was recorded every 1 to 2 minutes for 40 minutes using Bio-Tek KC4 v3.4 software.
Apoptosis Assay
HeLa cells were seeded into 12 wells of two 6-well plates (0.53×106 cells per well) at 37°C in 5% CO2 and grown overnight to 80% confluence. Cells in each well were transfected with the described plasmids using Lipofectamine LTX transfection agent. Cells in wells 1–2 of the 1st plate were each transfected with 1580ng of plasmid pcDNA3.1(+)-Grb7, while cells in wells 3–6 of the 1st plate were each transfected with 1194ng of empty vector plasmid pcDNA3.1(+). Cells in the 2nd plate were transfected identically to the cells in the 1st plate. Two days post-transfection cells were serum starved at 37°C in 5% CO2 overnight. Next, cells in wells 1–4 of the 1st plate were treated with 1µM of Staurosporine for 2.5 hrs, while cells in wells 1–4 of the 2nd plate were treated with 85µM Etoposide for 26 hrs. Cell lysates were prepared (100µL lysis buffer per well) at the indicated times and the endogenous Hax1 from normalized HeLa cell lysates in each well of two plates was detected and analyzed by Western Blot.
CellTiter-Glo Luminescent Cell Viability Assay
HeLa cells were seeded in 6 wells of a 6-well plate (0.53×106 cells per well) at 37°C and 5% CO2, and grown overnight to 80% confluence. Cells in each well were transfected with the described plasmids using Lipofectamine LTX transfection agent. Cells in well 1 were transfected with 2388ng of the plasmid pcDNA3.1(+) vector; Cells in well 2 were transfected with 1194ng pcDNA3.1(+) vector and 1580ng pcDNA3.1(+)-Grb7. Cells in well 3 were transfected with 1791ng pcDNA3.1(+) vector and 695ng pcDNA3.1(+)-Hax1. Cells in well 4 were transfected with 1580ng pcDNA3.1(+)-Grb7, 597ng pcDNA3.1(+) vector and 695ng pcDNA3.1(+)-Hax1. Cells in well 5 were transfected with 683ng pcDNA3.1(+)-ProCaspase3, 1194ng pcDNA3.1(+) vector and 695ng pcDNA3.1(+)-Hax1. Cells in well 6 were transfected with 683ng pcDNA3.1(+)-ProCaspase3, 1580ng pcDNA3.1(+)-Grb7 and 695ng pcDNA3.1(+)-Hax1. (The molar ratios of pcDNA3.1(+)-Grb7, pcDNA3.1(+)-Hax1, and pcDNA3.1(+)-ProCaspase3 were 2:1:1 respectively). Two days post-transfection cells from each well were detached by TrypLE Express according to the manufacturer’s protocol (Life Technologies), resuspended in DMEM medium lacking FBS, and subcultured in two white opaque-walled 96-well plates (6 wells per plate and 5000 cells per well) at 37°C and 5% CO2 and grown overnight. Any remaining cells were preserved at −80 °C for future Western Blot analyses. Next, for each plate at 37 °C and 5% CO2, cells in 3 wells were treated with 1 µM Staurosporine, while 3 wells were treated with DMSO as a negative control (Each transfection condition was shared by cells in 6 wells of each plate). After 1–2 hours incubation, cell viability assays was sequentially performed on cells from two 96-well plates according to the manufacturer’s protocol (Promega). Finally, luminescence was recorded using a TECAN GENios Multifunction fluorescence, absorbance and luminescence microplate reader with TECAN Magellan3 software (integration time: 1000 ms; gain setting: 150).
Results
in vitro binding assays indicate the purified Grb7-RAPH domains directly interact with the purified Hax1 isoform 1
In our previous research, the Grb7-RAPH domains were shown to interact with Hax1 by Yeast 2 Hybrid screening, and full length Grb7 was shown to interact with Hax1 by co-immunoprecipitation from HeLa mammalian cells (Siamakpour-Reihani et al. 2011). Still to be determined is whether this interaction is direct and exclusive, since it is conceivable binding could require assistance from other proteins as part of a complex. In order to confirm our previous assumption, purified recombinant protein domains and proteins were prepared and the relevant in vitro binding assays were performed as follows.
The interaction between the 6xHis-SUMO-Grb7-RAPH and Hax1-6xHis proteins was verified using immobilized SUMO antibody to capture the protein complex. Specifically, SUMO antibody conjugated resin was used to probe a mixture containing purified 6xHis-SUMO-Grb7-RAPH and Hax1-6xHis proteins. As seen in the Western Blot results in Figure 1-B, 6xHis-SUMO-Grb7-RAPH binds to Hax1-6xHis. Hax1 antibody detected Hax1-6xHis in elution buffers in the presence of 6xHis-SUMO-Grb7-RAPH (lanes 4–6), while no detectable Hax-6xHis can be found in elution buffers of the negative control (lanes 7–9). For the negative control, 6xHis-SUMO was incubated with Hax-6xHis and immobilized SUMO antibody. This result demonstrates the Grb7-RAPH domain construct alone is sufficient for binding to Hax1, and the interaction does not require the presence of intermediary proteins.
The combination of GST-RAPH and Hax1-6xHis, or GST-Hax1 and RAPH-6xHis proteins was not used here to verify the interaction between Grb7-RAPH domains and Hax1, since the GST protein was found to bind nonspecifically to the Grb7-RAPH domains (unpublished results).
in vitro assays indicate the dimerization state of Grb7 does not affect its ability to bind Hax1 isoform 1
The Grb7 protein exists primarily as a homodimer at micromolar concentrations (Porter et al. 2005). Dimerization is principally mediated through the Grb7 C-terminal SH2 domain, although a weak homodimerization reaction has also been suggested for the Grb7-RAPH domains (Depetris et al. 2009). A phenylalanine to arginine mutation (F511R) in the C-terminal helix of the Grb7-SH2 domain has been shown to largely block dimerization in both the Grb7-SH2 domain alone and the intact protein (Chu et al. 2010; Porter et al. 2005). Purified samples of this Grb7 mutant protein were used to monitor the effect of Grb7 dimerization state on its ability to bind with the Hax1 protein.
Equimolar amounts of purified Grb7-6xHis and Grb7 (F511R)-6xHis were separately incubated with purified Hax-6xHis protein and immobilized Grb7 antibody, and tested for protein-protein interactions by the previously described in vitro binding assay. Grb7 antibody (H-70) recognizes an epitope within the first N-terminal 70 residues of Grb7; therefore it is unlikely the F511R mutation will affect antibody binding. It is also expected the F511R mutation is sufficiently removed from the Grb7-RAPH domains to decrease any possible binding effects caused by altered Grb7 protein tertiary structure.
As can be seen from the Western Blot results in Figure 1-C (lanes 4, 5, 7, 8), both Grb7-6xHis and Grb7 (F511R)-6xHis bind to Hax-6xHis. Hax1 antibody detected Hax-6xHis in elution buffers under both conditions, while no detectable Hax-6xHis is observed in the elution fractions of the negative control (Hax1-6xHis protein incubated with immobilized Grb7 antibody). This result indicates the dimerization state of Grb7 does not affect its ability to bind to Hax1, by in vitro assay using purified proteins.
Grb7 and Hax1 isoform 1 may colocalize partially to mitochondria in SKBR3 cells upon growth factor stimulation
In previous studies, Grb7 has been observed at the cell membrane, in focal contacts, the cytosol, and nucleus (Chu et al. 2010; Han et al. 2000; Shen et al. 2002; Tsai et al. 2010); Hax1 has been located at the cytosolic membrane faces of mitochondria and the endoplasmic reticulum (ER), in P-bodies, the cytosol, and the nucleus (Grzybowska et al. 2013; Jeyaraju et al. 2009; Simmen 2011; Trebinska et al. 2010).
The SKBR3 human breast cancer cell line overexpresses the Grb7 protein (Pero et al. 2007; Pradip et al. 2013), but it does not express detectable Hax1 protein isoforms (by Western Blot analysis). pcDNA3.1(+)-Hax1 plasmids were transiently transfected into SKBR3 cells for expression of untagged Hax1 isoform 1, and protein localization was observed by immunofluorescence analysis.
One of the previously observed Hax1 cellular locations is the mitochondrion. Therefore the mitochondrial marker MitoTracker Deep Red FM dye was first used to investigate potential localization of Hax1 to these organelles.
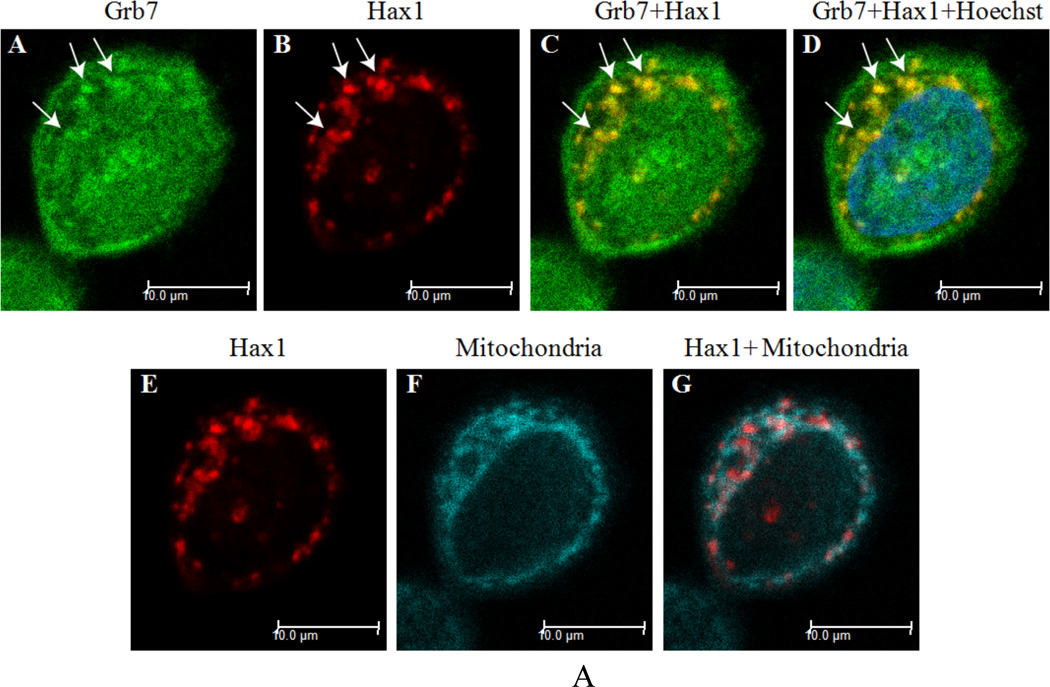
In Figure 2-A, Grb7 and Hax1 show apparent colocalization in SKBR3 cells grown in DMEM medium with 10% v/v FBS. Grb7 is visualized in Panels A, C, D; Hax1 is visualized in Panels B, C, D, E, G; mitochondria are visualized in Panels F and G; and the cell nucleus is visualized in Panel D. Areas of Grb7 and Hax1 overlap in Panels C and D visualize as yellow, with examples indicated by arrows. Although Grb7 and Hax1 obviously appear colocalized, their concurrence at mitochondrial locations is not readily apparent, as seen in panels E-G.
Figure 2. A) Grb7 and Hax1 isoform 1 colocalize in SKBR3 cells stained with mitochondria marker.
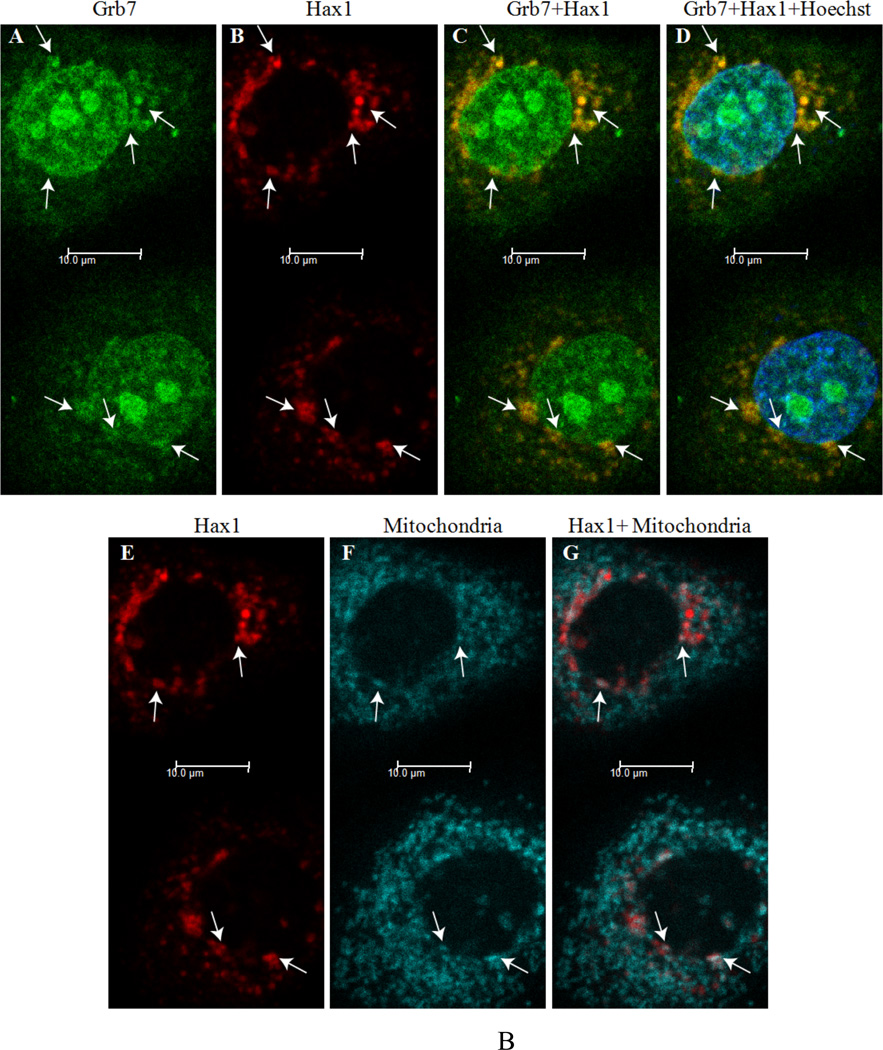
Two days post-transfection cells were stained with mitochondria marker. Colocalization sites of Grb7 and Hax1 are indicated by arrows and the overlapping yellow color in Panels C and D. Scale bar = 10 µm. B) Grb7 and Hax1 isoform 1 colocalize in EGF treated SKBR3 cells stained with mitochondria marker Two days post-transfection cells were serum starved, treated with EGF, and stained with mitochondria marker. Colocalization sites of Grb7 and Hax1 are indicated by arrows and the overlapping yellow color in Panels C and D. Possible colocalization sites with mitochondria are shown by arrows in Panel G. Scale bar = 10 µm.
Additionally, transfected SKBR3 cells were serum starved overnight and then treated with epidermal growth factor (EGF) to study the effect of growth factor stimulation on colocalization of Grb7 and Hax1. In Figure 2-B, Grb7 and Hax1 are again shown to colocalize in EGF treated SKBR3 cells, with several overlapping areas indicated by arrows in Panels C and D. Furthermore, by comparing the areas indicated by arrows in Panels E, F and G, there appear some areas of Hax1 localization with the mitochondria, with these areas overlapping the colocalization sites of Grb7 and Hax1 seen in Panels C and D. These results suggest EGF stimulation may induce the partial transfer of Grb7 and Hax1 to mitochondrial location. Alternatively, it is possible the Grb7 and Hax1 proteins could be localizing to an organelle or site not conclusively identified through these studies.
These results indicate Grb7 and Hax1 isoform 1 colocalize in SKBR3 cells, but their colocalization sites may reside on different organelles depending on cellular growth conditions. Specifically, Grb7 and Hax1 isoform 1 may colocalize partially to mithchondria induced by EGF stimulation.
To explore the possibility of Grb7/Hax1 colocalization in regions other than mitochondria, the intermediate- and trans- golgi marker Lectin GS-II Alexa Fluor 647 Conjugate was used (Supplemental Figures S1-A-C). SKBR3 cells were grown as untreated control, serum starved, and EGF stimulated to study protein colocalization. Again, Grb7 and Hax1 colocalize under all three growth conditions. It is possible there is some weak colocalization with the golgi marker in the serum starved cells, however, this evidence is tentative at best.
It is worth noting the specific identity of the cellular organelles or regions, to which Grb7 and Hax1 primarily colocalize remain unclear in SKBR3 cells. They appear to be larger than lysosomes, peroxisomes or P-bodies. It is possible they are endosomes, lysozyme polymers, or even golgi bodies, since the golgi probes used in these studies may not be sufficiently specific for this determination. Future studies in other cancer cell lines are required to better identify the exact identity of Grb7/Hax1 colocalization.
The Grb7 and Hax1 interaction can affect Caspase3 cleavage of Hax1 in vitro
Multiple previous studies have demonstrated Hax1 isoform 1 can inhibit apoptosis by interacting with different apoptosis-related proteins (Han et al. 2006; Lee et al. 2008; Vafiadaki et al. 2007; Vafiadaki et al. 2009). Additionally, at least one study has indicated Grb7 may suppress apoptosis but through an unclear mechanism (Giricz et al. 2012). Since Hax1 and Grb7 are both implicated in apoptosis, we wished to explore the possibility that Grb7 may indirectly regulate apoptosis via its interaction with Hax1. There is precedence for another member of the Grb7 family playing a role in apoptosis, specifically Grb10. Grb10 indirectly regulates apoptosis in HTC-IR cells (rat hepatoma cells overexpressing the insulin receptor) by binding to anti-apoptotic mitochondrial Raf1 kinase and MEK1 kinase (Han et al. 2001; Nantel et al. 1998; Nantel et al. 1999; Wang et al. 1996).
Caspase3, as well as Caspase9, is distributed in the mitochondria, cytosol, and nucleus (Chandra and Tang 2003; Kamada et al. 2005; Krajewski et al. 1999; Mancini et al. 1998; Ritter et al. 2000; Susin et al. 1999). Caspase9 is an initiator caspase involved in the activation cascade of caspases. Caspase3 is an effector caspase responsible for apoptosis execution (McIlwain et al. 2013) and is regarded as the primary caspase for degrading crucial regulatory and structural proteins (McIlwain et al. 2013; Slee et al. 2001). Hax1 is cleaved by Caspase3, while Caspase9 activation is inhibited by Hax1 (Han et al. 2006; Lee et al. 2008).
Taken together, we estimate Grb7 may indirectly affect apoptosis through its association with Hax1. Specifically, a Grb7/Hax1 association could interfere with subsequent Hax1 interactions with Caspases3 and 9, thus ultimately affecting the cell viability.
We focused on how Grb7 might affect the action of Caspase3 on Hax1 and propose a model for representing this process (Figure 3-A). In Figure 3-A top, a kinetic scheme for reversible enzyme inhibitors is presented. In our model (Figure 3-A top and bottom) the inhibitor (Grb7) combines with the free substrate (Hax1), and this combination may compete with binding between the enzyme and substrate. Based on this mechanism, Caspase3 as an enzyme cleaves free substrate Hax1, while Grb7 functions as an inhibitor through the formation of an SI complex that is resistant to Caspase3 cleavage. In Figure 3-B, the Caspase3 cleavage site in the Hax1 protein is indicated by a scissor emblem between residues Asp127 and Ser128 (Lee et al. 2008).
Figure 3. A) Top: Enzyme inhibition model for Caspase3 (enzyme), Hax1 (substrate) and Grb7 (inhibitor).
In the proposed model Grb7 (indicated by the circled “I” letter) acts as a substrate inhibitor as described in the text. Bottom: Model of Grb7 inhibition in Caspase3 cleavage of Hax1 Grb7 as an inhibitor binds to Hax1, affecting Caspase3 cleavage of Hax1. B) Caspase3 cleavage site on Hax1 Caspase3 cleaves between D127 and S128 of Hax1, producing the 14kDa and 19kDa cleavage products discussed in the text.
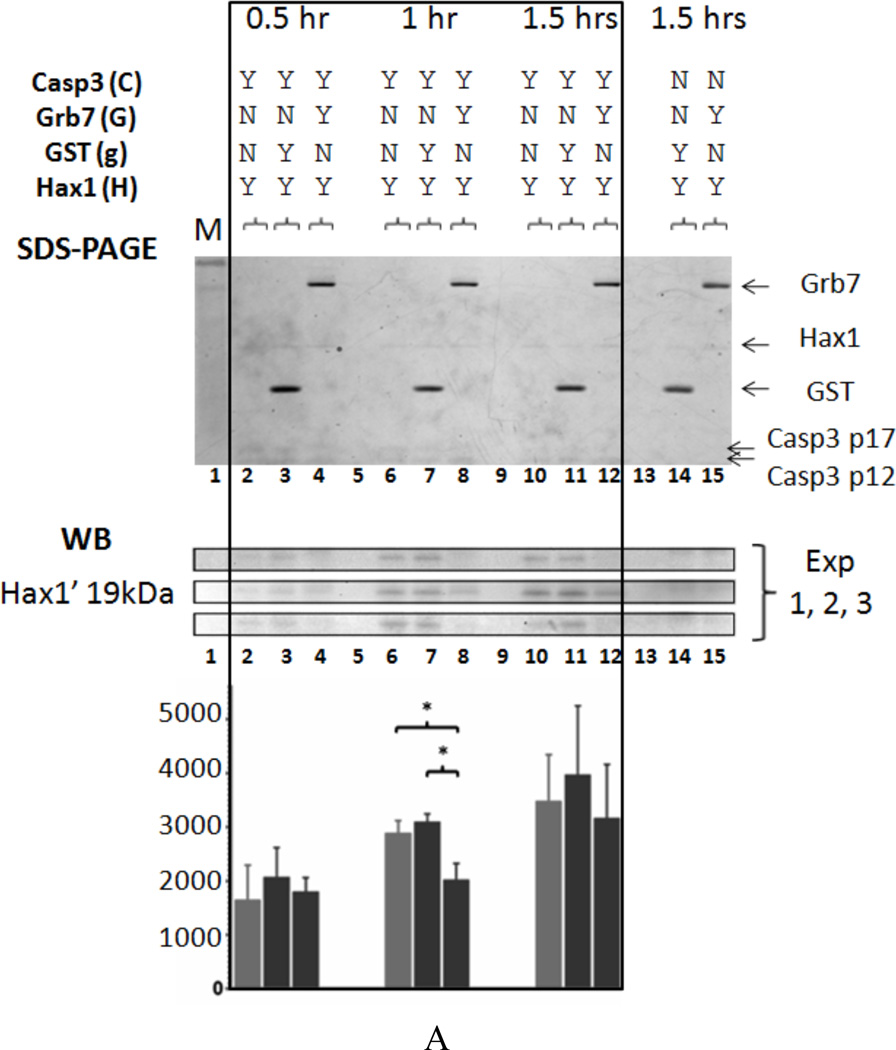
To investigate the role of Grb7 in the proposed model, an in vitro Caspase3 cleavage of Hax1 assay was performed. Purified recombinant Caspase3-6xHis, Grb7-6xHis and Hax1-6xHis proteins were incubated together in a molar ratio of 1:10:1 (respectively) at 37°C. Samples were collected every 30 minutes to a final time period of 1.5 hours. The samples were then probed with anti-Hax1 antibody, analyzed by Western Blot and the results represented in Figure 4-A. An equivalent amount of GST was used to replace Grb7-6xHis (i.e. reagent molar ratios again at 1:10:1) to rule out the possibility that Grb7 affects the cleavage of Hax1 through nonspecific protein crowding. In Figure 4-A (Coomassie Blue gel results) Grb7-6xHis can be seen in lanes 4, 8, 12, and 15, while GST appears in lanes 3, 7, 11, and 14. Intact Hax-6xHis (lanes 2–4, 6–8, 10–12, 14–15) and Caspase3 subunits p17 (17kDa) and p12 (12kDa) (lanes 2–4, 6–8, 10–12) appear at weaker intensity, presumably due to their lower relative concentrations. In Figure 4-A (Western Blot results), Hax1 cleavage product Hax1’ (19kDa) is seen in both Caspase3 and Hax1 (CH), and Caspase3, GST and Hax1 (CgH) cases at three time points: 0.5 hr, 1 hr and 1.5 hrs. However, in the Caspase3, Grb7 and Hax1 case (CGH), the cleavage products are observed with weaker intensity than in the CH and CgH cases. This is especially apparent at the 1 hr time point. This assay was repeated three times. The densitometric analysis of the Hax1’ cleavage product signal for the CGH case at the 1 hr time point indicates the presence of significantly less cleavage product than in the CgH and CH cases. However, this effect is not as obvious at the 0.5 hr and 1 hr time points. The abnormally weak Hax1’ signal in lane 10 of the 3rd experiment could be caused by an error in protein migration during electrophoresis. Taken together, these results indicate Grb7 is capable of inhibiting Caspase3 cleavage of Hax1, possibly in a time dependent manner.
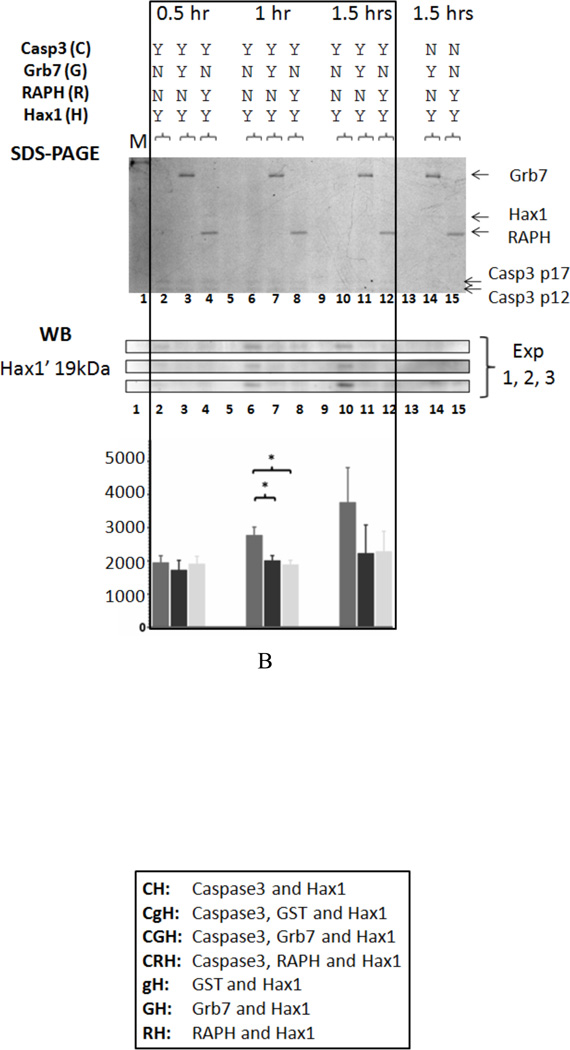
Figure 4. A) Coomassie Blue staining, Western Blot, and densitometric analysis results for the in vitro Caspase3 cleavage assay I.
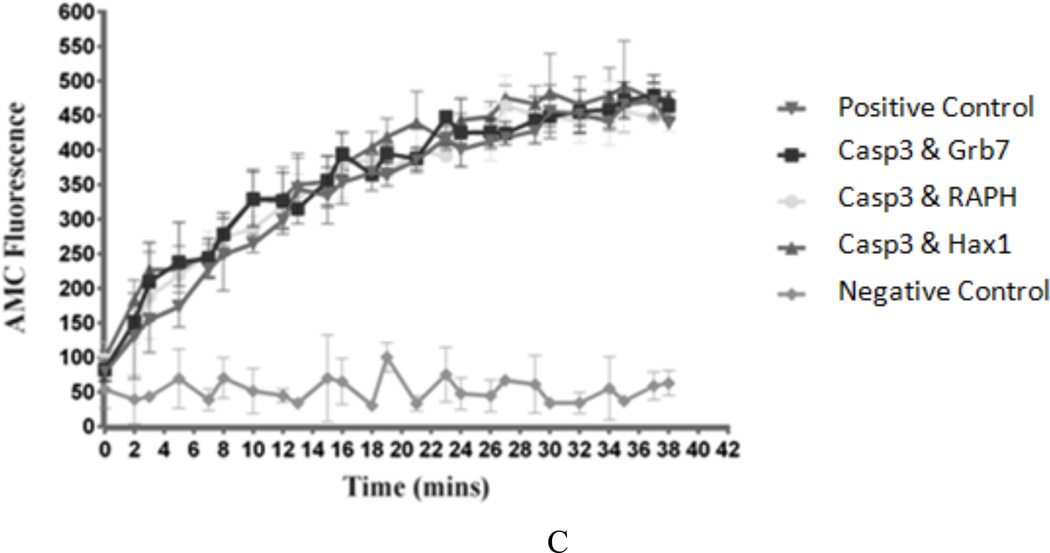
Lane 1: Protein Marker. Lane 2: CH: Caspase3 and Hax1 at 0.5 hr. Lane 3: CgH: Caspase3, GST and Hax1 at 0.5 hr. Lane 4: CGH: Caspase3, Grb7 and Hax1 at 0.5 hr. Lane 5: Blank. Lane 6: CH at 1 hr. Lane 7: CgH at 1 hr. Lane 8: CGH at 1 hr. Lane 9: Blank. Lane 10: CH at 1.5 hrs. Lane 11: CgH at 1.5 hrs. Lane 12: CGH at 1.5 hrs. Lane 13: Blank. Lane 14: gH: GST and Hax1 at 1.5 hrs. Lane 15: GH: Grb7 and Hax1 at 1.5 hrs. The assay was repeated three times and the amounts of cleavage product Hax1’ (19kDa) were studied by Western Blot and densitometric analysis. Asterisks indicate statistically relevant results (P values < 0.02) B) Coomassie Blue staining, Western Blot and densitometric analysis results for the in vitro Caspase3 cleavage assay II Lane 1: Protein Marker. Lane 2: CH: Caspase3 and Hax1 at 0.5 hr. Lane 3: CGH: Caspase3, Grb7 and Hax1 at 0.5 hr. Lane 4: CRH: Caspase3, RAPH and Hax1 at 0.5 hr. Lane 5: Blank. Lane 6: CH at 1 hr. Lane 7: CGH at 1 hr. Lane 8: CRH at 1 hr. Lane 9: Blank. Lane 10: CH at 1.5 hrs. Lane 11: CGH at 1.5 hrs. Lane 12: CRH at 1.5 hrs. Lane 13: Blank. Lane 14: GH: Grb7 and Hax1 at 1.5 hrs. Lane 15: RH: RAPH and Hax1 at 1.5 hrs. The assay was repeated three times and the cleavage product Hax1’ (19kDa) was analyzed by Western Blot and densitometric analysis. Asterisks indicate statistically relevant results (P values < 0.02). C) in vitro Caspase3 activity assay Five different cases are shown: Ac-DEVD-AMC only (negative control), Caspase3 with Ac-DEVD-AMC (positive control), and Grb7-6xHis, RAPH-6xHis or Hax1-6xHis incubated with Caspase3 and Ac-DEVD-AMC. The fluorescence signal from released AMC was recorded every 1–2 minutes for a total of 40 minutes.
Next, we investigated whether the Grb7-RAPH domains alone could affect Caspase3 cleavage of Hax1. The molar ratio of Caspase3-6xHis, Grb7-6xHis (or Grb7-RAPH-6xHis) and Hax1-6xHis to each other was 1:10:1, as previously, and samples were collected every 30 minutes for a final time of 1.5 hours. In Figure 4-B (Coomassie Blue gel results), Grb7-6xHis can be seen in lanes 3, 7, 11, and 14, while Grb7-RAPH-6xHis appears in lanes 4, 8, 12, and 15. Intact Hax1-6xHis (lanes 2–4, 6–8, 10–12, 14–15) and Caspase3 subunits p17 (17kDa) and p12 (12kDa) (lanes 2–4, 6–8, 10–12) appear at weaker intensity presumably due to their lower relative concentrations. In Figure 4-B (Western Blot results), Hax1 cleavage products Hax1’ (19kDa) is seen in the Caspase3 and Hax1 (CH) case at all three time points: 0.5 hr, 1 hr and 1.5 hrs. However, for both the Caspase3, Grb7 and Hax1 (CGH), and the Caspase3, Grb7-RAPH and Hax1 (CRH) cases, the Hax1 cleavage products are observed with weaker intensity than in the CH case. This is especially apparent at the 1 hr time point. This assay was also repeated three times. The densitometric analysis results indicate the Grb7-RAPH domains alone can inhibit Caspase3 cleavage of Hax1, similarly to full length Grb7. They also demonstrate inhibition could potentially be time dependent.
It is possible Grb7 or the Grb7-RAPH domains may interact directly with Caspase3 to alter its activity, rather than inhibiting the cleavage reaction between Caspase3 and Hax1. To rule out this possibility, a common substrate to measure Caspase3 activity (Ac-DEVD-AMC) was used to measure Caspase3 activity in the presence of Grb7 and the Grb7-RAPH domains. For this assay, Grb7-6xHis, Grb7-RAPH-6xHis, or Hax1-6xHis proteins were separately incubated with Caspase3-6xHis and Ac-DEVD-AMC (Figure 4-C) for quantifying Caspase3 activity. The molar ratio of Grb7-6xHis (or Grb7-RAPH-6xHis, or Hax1-6xHis) and Caspase3-6xHis to each other was 10:1.
In Figure 4-C, the release of fluorogenic AMC upon cleavage of Ac-DEVD-AMC provides a quantitative assessment of Caspase3 activity. It can be observed in all three cases: Caspase3 with Grb7, Caspase3 with Grb7-RAPH and Caspase3 with Hax1, the emitted AMC fluorescence is indistinguishable from the fluorescence signal of the positive control over at least a 40 minute time period. This result suggests Caspase3 innate activity is not affected by the presence of Grb7, the Grb7-RAPH domains, or Hax1 in vitro.
It is noted the fluorescence signal from the Ac-DEVD-AMC cleaved peptide for the Caspase3/Hax1 combination is not noticeably decreased, nor displays a time lag, in comparison to the Grb7 and Grb7-RAPH cases. This is despite the fact we, and others (Lee et al. 2008), argue Hax1 is a substrate for Caspase3. One would expect to see an effect due to the presence of two potentially competing substrates, Ac-DEVD-AMC and Hax1. One possibility exists for the lack of any observable effect of Hax1 on Caspase3 cleavage of Ac-DEVD-AMC. The Ac-DEVD-AMC peptide contains the optimal Caspase3 DEVD cleavage site, while the Caspase3 Hax1 cleavage site consists of TLRD. It could be Caspase3 is relatively unreactive towards Hax1 in the presence of a more optimal substrate.
In sum, our results support the theory that Grb7 can inhibit Caspase3 cleavage of Hax1 isoform 1 in a potentially time dependent manner in vitro. This inhibition is achieved by the interaction between Hax1 and Grb7 through the Grb7-RAPH domains, and Grb7 does not interact directly with Caspase3 to affect its innate activity. Our studies support an inhibitory role of Grb7 on Caspase3 cleavage by interfering with the association of Caspase3 and Hax1.
Grb7 expression may slow Caspase3 cleavage of Hax1 isoform 1 in apoptotic HeLa cells
Apoptosis assays were performed to determine the role of Grb7 in Hax1 protein integrity during apoptosis. The HeLa cell line was chosen for these assays because it is a commonly used apoptosis model. HeLa cells mainly express endogenous Hax1 isoform 1 (Grzybowska et al. 2013; Jeyaraju et al. 2009), and they also express endogenous ProCaspase3 (Laussmann et al. 2011). HeLa cells do not express detectable Grb7 isoform 1 (by Western Blot analysis), Grb7 isoform 1 is the most common isoform (at 532 aa) of the human Grb7 protein (Tanaka et al. 1998).
The small molecule drugs Staurosporine (STS) and Etoposide (ETO) both cause cellular apoptosis: STS inhibits protein kinases while ETO interrupts DNA synthesis (Chae et al. 2000; Day et al. 2009; Hande 1998; Karaman et al. 2008). Hence these two drugs were utilized to induce apoptosis in HeLa cells and to study the effect of Grb7 expression on Hax1 protein integrity during apoptosis.
For the assay, pcDNA3.1(+)-Grb7 plasmids were transfected into HeLa cells for expression of untagged Grb7 protein. Two days post-transfection cells were serum-starved overnight, followed by treatment with 1µM STS for 2.5 hours or 85µM ETO for 26 hours. Cell lysates were prepared at the indicated times and the endogenous Hax1 levels were detected by Western Blot (Figure 5).
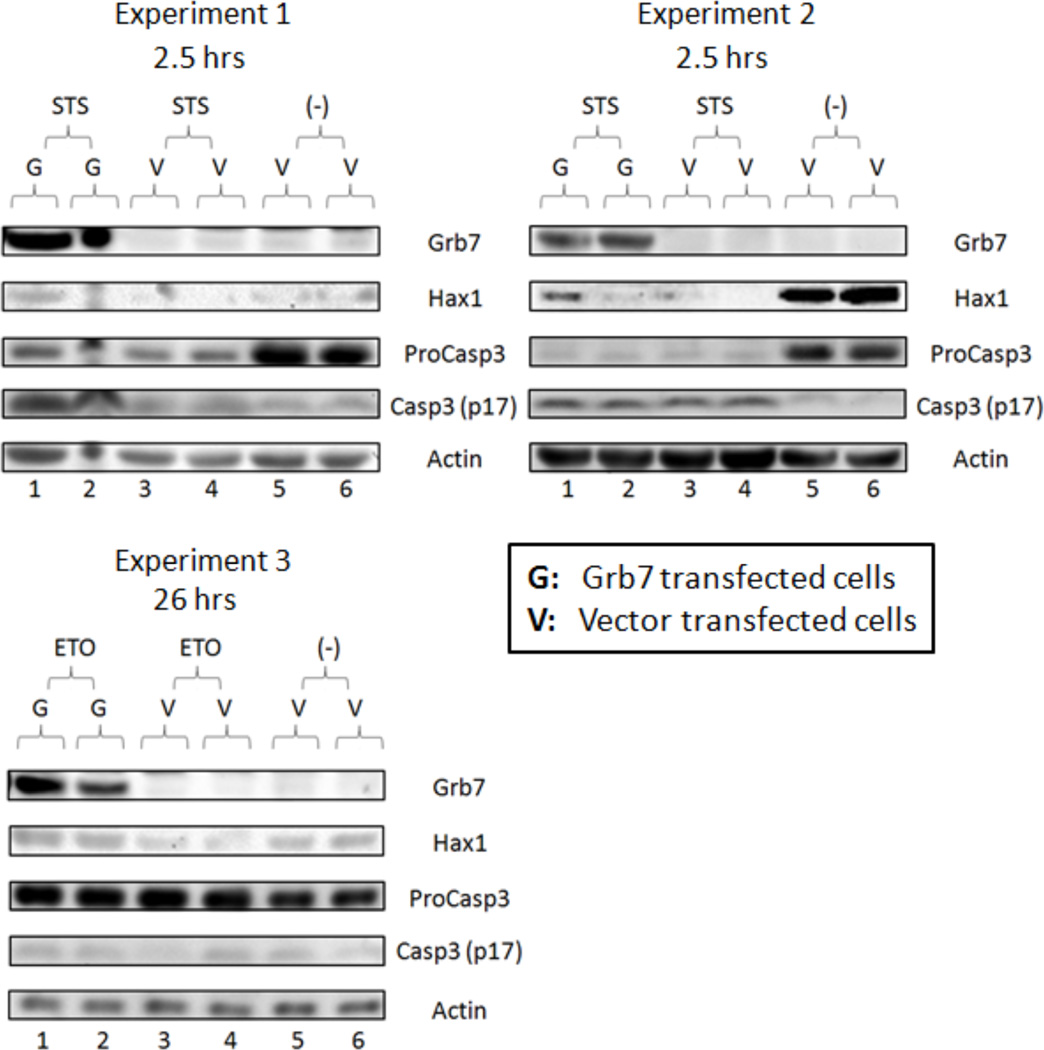
Figure 5. Grb7 may slow cleavage or degradation of Hax1 isoform 1 in apoptotic HeLa cells. Top (Experiments 1 and 2): Western Blot results for two STS induced apoptosis trials in HeLa cells.
Lane 1–2: cell lysates from apoptotic Grb7 transfected cells. Lane 3–4: cell lysates from apoptotic vector transfected cells. Lane 5–6: untreated vector transfected cell lysates. Bottom (Experiment 3): Western Blot results for a single ETO induced apoptosis trial in HeLa cells Lane 1–2: cell lysates from apoptotic Grb7 transfected cells. Lane 3–4: cell lysates from apoptotic vector transfected cells. Lane 5–6: untreated vector transfected cell lysates.
In HeLa cells treated with STS (Figure 5 top lanes 3–4 in both gels), an appreciable decrease in Hax1 signal is noted relative to the Hax1 signal observed in non-drug treated cells (Figure 5 top lanes 5–6 in both gels). This decrease is likely due to accumulated active Caspase3 over time after STS treatment. We base this conclusion upon the observation that active Caspase3 signals are stronger in lane 3–4 than lane 5–6, and Procaspase3 signals are weaker in lane 3–4 than lane 5–6 in both gels (Figure 5 top). In Grb7 transfected and STS treated cells (Figure 5 top lanes 1–2 in both gels) the Hax1 signals are clearly stronger relative to drug treated cells that are not Grb7 transfected (Figure 5 top lanes 3–4 in both gels). The abnormally weak Hax1 signal in lane 2 of the 2nd experiment is likely caused by an error in protein migration during electrophoresis.
In HeLa cells treated with ETO (Figure 5 bottom lanes 3–4), a decrease in the Hax1 signal is also noted relative to the Hax1 signal observed in non-drug treated cells (Figure 5 bottom lanes 5–6). This decrease could be achieved through proteasomic degradation or other unknown processes, since the active Caspase3 signals are weak in all lanes and Procaspase3 signals are similar in all lanes (Figure 5 bottom). In Grb7 transfected and ETO treated cells (Figure 5 bottom lanes 1–2) the Hax1 signals are clearly stronger relative to drug treated cells that are not Grb7 transfected (Figure 5 bottom lanes 3–4).
The result indicates Grb7 can slow Hax1 isoform 1 cleavage and/or degradation in apoptotic HeLa cells. This effect may not be confined to Grb7-mediated resistance to Caspase3 cleavage, but could also be due to proteasomic or other unknown degradation processes.
Grb7 expression may increase cell viability in apoptotic HeLa cells in a time dependent manner
Cell viability assays were performed to determine the role of Grb7 in cell viability during apoptosis. In these assays, co-transfection and triple-transfection were performed in order to discover how Grb7 and Hax1 expression could affect cell viability in the absence or presence of overexpressed Caspase3.
Two days post-transfection cells of each group were serum-starved overnight while remaining cells were harvested for Western Blot analysis. Following starvation, cells were treated with 1µM STS for 1.5 and 2.5 hours. CellTiter-Glo luminescent cell viability assays were sequentially performed on cells at the two time points.
In Figure 6 top, Grb7 expression is evident in Groups 2, 4, and 6. However, Hax1 (or ProCaspase3) overexpression is not apparent in Groups 3–6 (or Groups 5–6 for ProCaspase3). In these transfected cells, the Hax1 and ProCaspase3 signals are not appreciably stronger than the endogenous Hax1 and ProCaspase3 signals seen in the Hax1 (or ProCaspase3) non-transfected Groups 1 (V) and 2 (VG). This lack of apparent Hax1 and ProCaspase3 transfection efficiency could be due to the molar ratios of the plasmids for transfection (The molar ratios of pcDNA3.1(+)-Grb7, pcDNA3.1(+)-Hax1, and pcDNA3.1(+)-ProCaspase3 were 2:1:1 respectively). In addition, from Figure 6 top, Grb7 expression does not up-regulate the expression of endogenous Hax1 before drug treatment. This discovery implies Grb7 may affect Hax1 cleavage/degradation in apoptotic HeLa cells through its physical association with endogenous Hax1 rather than up-regulation of Hax1 expression.
Figure 6. Top: Western Blot results for transfected cells prior to STS induced apoptosis.
Lane 1–6: Lysates from HeLa cells transfected with different sets of plasmids. V: pcDNA3.1(+) vector, G: pcDNA3.1(+)-Grb7, H: pcDNA3.1(+)-Hax1, C: pcDNA3.1(+)-ProCaspase3. Bottom: CellTiter-Glo luminescent cell viability assay for transfected cells after STS induced apoptosis Viability ratios for all protein expression combinations (indicated in the figure legend) are graphically reported at the 1.5 hrs and 2.5 hrs time points. Asterisks indicate statistically relevant results (P values < 0.02).
In Figure 6 bottom, the luminescence ratio of STS treated cells to untreated cells is used to evaluate cell viability (viability ratio). At the 1.5 hrs time point cells of every group have a similar viability ratio of approximately 0.8. At the 2.5 hrs time point the viability ratio of all groups dropped by almost 50%. Furthermore, at this time point the viability ratio of Group 4 (GVH) is significantly greater than Group 3 (VH), and the viability ratio of Group 6 (CGH) is significantly greater than Group 5 (CVH). The viability ratio of Group 2 (VG) is slightly greater than Group 1 (V), however, this difference is not statistically significant. These results suggest expression of Grb7 may increase cell viability in STS induced apoptotic HeLa cells in a time dependent manner. Further studies are needed to clarify this potential role of Grb7 expression in anti-apoptotic signaling pathways.
Discussion
Through these studies we have shown the signal transducing adaptor protein Grb7 can exclusively bind to the anti-apoptotic protein Hax1 isoform 1 in vitro through its RA and PH domains, and the interaction does not depend on Grb7 protein dimerization state. Additionally, via confocal immunofluorescence analysis, we report Grb7 and Hax1 isoform 1 may colocalize partially to mitochondria in SKBR3 cells treated with EGF. We have demonstrated Grb7 can inhibit Caspase3 cleavage of Hax1 in a time dependent manner in vitro, and Grb7 expression may slow cleavage or degradation of endogenous Hax1 isoform 1 in apoptotic HeLa cells induced by STS and ETO. Our results have also indicated Grb7 protein expression may increase cell viability in STS induced apoptotic HeLa cells in a time dependent manner. In our prior study (Siamakpour-Reihani et al. 2011), Hax1 was found to interact with the Grb7-RAPH domains using Yeast 2 Hybrid screening methods. The Grb7-RAPH domains were used alone as bait in screening since tandem RAPH domains are known to act as functional protein units in some cases (Depetris et al. 2009). As well, it is possible that non-mammalian expression of mammalian proteins may affect the accessibility of protein domains for subsequent signaling events due to differences in post-translational modifications. Our previous co-immunoprecipitation assays from HeLa cell lysates suggested mutation of tyrosine residues in the Grb7 protein could hinder its ability to bind to Hax1 (Siamakpour-Reihani et al. 2011). However, our current study has shown the Grb7-RAPH domains alone are sufficient for interaction with Hax1 by in vitro binding assays. Since all recombinant proteins were expressed and purified from E.coli bacteria and presumably lack tyrosine phosphorylation or other modifications, our results suggest Grb7 is capable of binding to Hax1 in a non-mammalian environment. As well, our investigation demonstrated a known Grb7 mutation F511R that abrogates dimerization (Porter et al. 2005) does not affect the Grb7/Hax1 interaction. Therefore, according to these results, post-translational protein modifications and/or dimerization state may not affect Grb7 signaling through Hax1. This result does not preclude the possibility that tyrosine phosphorylated Grb7 in human cancer cells may interact with Hax1 more strongly than Grb7 recombinant protein in in vitro assays.
It would be desirable to know the strength of in vitro binding between Hax1 and Grb7 in a quantitative sense. Future experiments will focus on achieving this, and may include ELISA-based experiments, or immunoprecipitation experiments performed over a range of molar concentrations. Additionally, measurement of the binding kinetics of the Grb7:Hax1 interaction (e.g. by SPR) could help to explain the relatively long time-dependence observed for the inhibition of caspase3 cleavage of Hax1 by Grb7.
We have demonstrated, Grb7 and Hax1 isoform 1 colocalize strongly in SKBR3 cells, and Grb7/Hax1 colocalization may involve more than one type of organelle or cellular body. Our studies indicate partial Grb7/Hax1 colocalization to mitochondria in EGF treated SKBR3 cells; or possibly some colocalization to golgi in serum starved SKBR3 cells (supplemental results), although Grb7/Hax1 golgi colocalization appears weak at best. The immunofluorescence confocal images in this study indicate many locations of Grb7/Hax1 colocalization in SKBR3 cells remain inconclusively identified. Interestingly, Zayat et al. (2015) also noted the presence of unidentified granular bodies containing Hax1 that were not P-bodies (Zayat et al. 2015). In that study, HeLa cells treated with puromycin, and primary neuronal cells, both displayed two types of Hax1 cytoplasmic granules. These were Hax1 and DCP1-containing P-bodies (DCP1 is a P-body marker protein), and granules containing HAX1 but not DCP1. Future work will involve the use of different and/or more specific organelle markers, or colloidal gold-labeled cells, to further investigate the identity of these areas of Grb7/Hax1 cellular colocalization. It is also possible the overexpression of recombinant Hax-1 protein in SKBR3 cells can affect its cellular localization in a non-physiologically relevant manner. Therefore, it will be important in the future to investigate Grb7/Hax1 colocalization in cells that endogenously express both Hax1 and Grb7. Examples of cell lines that could perhaps be utilized for this purpose include A431, an epithelial carcinoma cell line, or HepG2, a liver carcinoma cell line.
We have shown Grb7 can alter Caspase3 cleavage of Hax1 in a time dependent manner in vitro, and its expression may also slow endogenous Hax1 isoform 1 cleavage or degradation in apoptotic HeLa cells. The in vitro assay and apoptosis assay on HeLa cells complement each other, with both implying possible functional consequences of the Grb7/Hax1 interaction. In unpublished results, we also observe considerable Grb7 degradation in apoptotic HeLa cells. Even under this adverse condition, the remaining intact Grb7 proteins are still capable of slowing the Hax1 cleavage process (Figure 5).
In cell viability assays, expression of Grb7 results in a measurable increase in cell viability in STS induced HeLa cells at the 2.5 hrs exposure time point (Figure 6 bottom). A Grb7 expression effect on cell viability is not readily apparent at the 1.5 hrs time point, and may simply reflect a lack of sufficient apoptosis progress. With multiple binding partners in several signaling pathways, the influence of Grb7 on apoptosis and viability may not be restricted to its signaling through Hax1, but could be augmented by other poorly defined (at present) pathways. The relative importance of Grb7 signaling via Hax1 in apoptosis processes remains to be fully defined.
Conclusions
Taken together, our studies have provided a direct link between the signal adapter protein Grb7, anti-apoptotic protein Hax1 isoform 1, and Caspase3 mediated apoptosis pathways. They suggest that as well as modulating cell migration signal transduction, Grb7 may also participate in Hax1 related apoptosis pathways mediated by Caspase3. We propose that Grb7’s demonstrated anti-apoptotic effect (Giricz et al. 2012; Sahlberg et al. 2013) may be achieved through its interaction with Hax1 isoform 1. Further, through our studies expression of Grb7 is shown to slow Hax1 cleavage/degradation and increase viability in apoptotic HeLa cells. We readily acknowledge further investigation is warranted to determine whether the Grb7/Hax1 interaction is a major route for apoptosis escape in Grb7 overexpressing tumors, or simply a minor pathway of Grb7 signaling.
Supplementary Material
Acknowledgments
We thank Dr. Jeongwon Jun for DNA sequencing support, Drs. Aaron Rowland and Kevin Houston for tissue culture support, and Drs. Erik Yukl and Brad Shuster for valuable discussions.
Abbreviations
- AMC
7-Amino-4-Methyl Coumarin
- Caspase3
Cysteine dependent aspartate directed protease 3
- ErbB2
Erythroblastosis oncogene B 2
- Grb7
Growth factor receptor bound protein 7
- GST
Glutathione S-Transferase
- Hax1
HS1 associated protein X1 protein
- HER2
Human Epidermal Growth Factor Receptor 2
- IP
Immunoprecipitation
- PH
Pleckstrin Homology
- RA
Ras Associating
- SH2
Src Homology 2
References
- Cavnar PJ, Berthier E, Beebe DJ, Huttenlocher A. Hax1 regulates neutrophil adhesion and motility through RhoA. J Cell Biol. 2011;193(3):465–473. doi: 10.1083/jcb.201010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HJ, Kang JS, Byun JO, Han KS, Kim DU, Oh SM, Kim HM, Chae SW, Kim HR. Molecular mechanism of staurosporine-induced apoptosis in osteoblasts. Pharmacol Res. 2000;42(4):373–381. doi: 10.1006/phrs.2000.0700. [DOI] [PubMed] [Google Scholar]
- Chandra D, Tang DG. Mitochondrially localized active caspase-9 and caspase-3 result mostly from translocation from the cytosol and partly from caspase-mediated activation in the organelle. Lack of evidence for Apaf-1-mediated procaspase-9 activation in the mitochondria. J Biol Chem. 2003;278(19):17408–17420. doi: 10.1074/jbc.M300750200. [DOI] [PubMed] [Google Scholar]
- Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature. 2008;452(7183):98–102. doi: 10.1038/nature06604. [DOI] [PubMed] [Google Scholar]
- Chu PY, Li TK, Ding ST, Lai IR, Shen TL. EGF-induced Grb7 recruits and promotes Ras activity essential for the tumorigenicity of Sk-Br3 breast cancer cells. J Biol Chem. 2010;285(38):29279–29285. doi: 10.1074/jbc.C110.114124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PY, Huang LY, Hsu CH, Liang CC, Guan JL, Hung TH, Shen TL. Tyrosine phosphorylation of growth factor receptor-bound protein-7 by focal adhesion kinase in the regulation of cell migration, proliferation, and tumorigenesis. J Biol Chem. 2009;284(30):20215–20226. doi: 10.1074/jbc.M109.018259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilenti L, Soundarapandian MM, Kyriazis GA, Stratico V, Singh S, Gupta S, Bonventre JV, Alnemri ES, Zervos AS. Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2 protease during cell death. J Biol Chem. 2004;279(48):50295–50301. doi: 10.1074/jbc.M406006200. [DOI] [PubMed] [Google Scholar]
- Daly RJ, Sanderson GM, Janes PW, Sutherland RL. Cloning and characterization of GRB14, a novel member of the GRB7 gene family. J Biol Chem. 1996;271(21):12502–12510. doi: 10.1074/jbc.271.21.12502. DOI. [DOI] [PubMed] [Google Scholar]
- Daly RJ. The Grb7 family of signalling proteins. Cell Signal. 1998;10(9):613–618. doi: 10.1016/s0898-6568(98)00022-9. [DOI] [PubMed] [Google Scholar]
- Day TW, Wu CH, Safa AR. Etoposide induces protein kinase Cdelta- and caspase-3-dependent apoptosis in neuroblastoma cancer cells. Mol Pharmacol. 2009;76(3):632–640. doi: 10.1124/mol.109.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denault JB, Salvesen GS. Expression, purification, and characterization of caspases. Curr Protoc Protein Sci. 2003 doi: 10.1002/0471140864.ps2113s30. Chapter 21:Unit 21.13. [DOI] [PubMed] [Google Scholar]
- Depetris RS, Wu J, Hubbard SR. Structural and functional studies of the Ras-associating and pleckstrin-homology domains of Grb10 and Grb14. Nat Struct Mol Biol. 2009;16(8):833–839. doi: 10.1038/nsmb.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeel B, Grzybowska E. HAX-1: a multifunctional protein with emerging roles in human disease. Biochim Biophys Acta. 2009;1790(10):1139–1148. doi: 10.1016/j.bbagen.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Fiddes RJ, Campbell DH, Janes PW, Sivertsen SP, Sasaki H, Wallasch C, Daly RJ. Analysis of Grb7 recruitment by heregulin-activated erbB receptors reveals a novel target selectivity for erbB3. J Biol Chem. 1998;273(13):7717–7724. doi: 10.1074/jbc.273.13.7717. [DOI] [PubMed] [Google Scholar]
- Frantz JD, Giorgetti-Peraldi S, Ottinger EA, Shoelson SE. Human GRB-IRbeta/GRB10. Splice variants of an insulin and growth factor receptor-binding protein with PH and SH2 domains. J Biol Chem. 1997;272(5):2659–2667. doi: 10.1074/jbc.272.5.2659. [DOI] [PubMed] [Google Scholar]
- Giricz O, Calvo V, Pero SC, Krag DN, Sparano JA, Kenny PA. GRB7 is required for triple-negative breast cancer cell invasion and survival. Breast Cancer Res Treat. 2012;133(2):607–615. doi: 10.1007/s10549-011-1822-6. [DOI] [PubMed] [Google Scholar]
- Grzybowska EA, Zayat V, Konopiński R, Trębińska A, Szwarc M, Sarnowska E, Macech E, Korczyński J, Knapp A, Siedlecki JA. HAX-1 is a nucleocytoplasmic shuttling protein with a possible role in mRNA processing. FEBS J. 2013;280(1):256–272. doi: 10.1111/febs.12066. [DOI] [PubMed] [Google Scholar]
- Han DC, Guan JL. Association of focal adhesion kinase with Grb7 and its role in cell migration. J Biol Chem. 1999;274(34):24425–24430. doi: 10.1074/jbc.274.34.24425. [DOI] [PubMed] [Google Scholar]
- Han DC, Shen TL, Guan JL. The Grb7 family proteins: structure, interactions with other signaling molecules and potential cellular functions. Oncogene. 2001;20(44):6315–6321. doi: 10.1038/sj.onc.1204775. [DOI] [PubMed] [Google Scholar]
- Han DC, Shen TL, Guan JL. Role of Grb7 targeting to focal contacts and its phosphorylation by focal adhesion kinase in regulation of cell migration. J Biol Chem. 2000;275(37):28911–28917. doi: 10.1074/jbc.M001997200. [DOI] [PubMed] [Google Scholar]
- Han Y, Chen YS, Liu Z, Bodyak N, Rigor D, Bisping E, Pu WT, Kang PM. Overexpression of HAX-1 protects cardiac myocytes from apoptosis through caspase-9 inhibition. Circ Res. 2006;99(4):415–423. doi: 10.1161/01.RES.0000237387.05259.a5. [DOI] [PubMed] [Google Scholar]
- Hande KR. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur J Cancer. 1998;34(10):1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Siddle K. Grb10 and Grb14: enigmatic regulators of insulin action--and more? Biochem J. 2005;388(Pt 2):393–406. doi: 10.1042/BJ20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaraju DV, Cisbani G, De Brito OM, Koonin EV, Pellegrini L. Hax1 lacks BH modules and is peripherally associated to heavy membranes: implications for Omi/HtrA2 and PARL activity in the regulation of mitochondrial stress and apoptosis. Cell Death Differ. 2009;16(12):1622–1629. doi: 10.1038/cdd.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada S, Kikkawa U, Tsujimoto Y, Hunter T. Nuclear translocation of caspase-3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s) J Biol Chem. 2005;280(2):857–860. doi: 10.1074/jbc.C400538200. [DOI] [PubMed] [Google Scholar]
- Kang YJ, Jang M, Park YK, Kang S, Bae KH, Cho S, Lee CK, Park BC, Chi SW, Park SG. Molecular interaction between HAX-1 and XIAP inhibits apoptosis. Biochem Biophys Res Commun. 2010;393(4):794–799. doi: 10.1016/j.bbrc.2010.02.084. [DOI] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Kasus-Jacobi A, Béréziat V, Perdereau D, Girard J, Burnol AF. Evidence for an interaction between the insulin receptor and Grb7. A role for two of its binding domains, PIR and SH2. Oncogene. 2000;19(16):2052–2059. doi: 10.1038/sj.onc.1203469. [DOI] [PubMed] [Google Scholar]
- Koontz J, Kontrogianni-Konstantopoulos A. Competition through dimerization between antiapoptotic and proapoptotic HS-1-associated protein X-1 (Hax-1) J Biol Chem. 2014;289(6):3468–3477. doi: 10.1074/jbc.M113.536151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski S, Krajewska M, Ellerby LM, Welsh K, Xie Z, Deveraux QL, Salvesen GS, Bredesen DE, Rosenthal RE, Fiskum G, Reed JC. Release of caspase-9 from mitochondria during neuronal apoptosis and cerebral ischemia. Proc Natl Acad Sci USA. 1999;96(10):5752–5757. doi: 10.1073/pnas.96.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laussmann MA, Passante E, Düssmann H, Rauen JA, Würstle ML, Delgado ME, Devocelle M, Prehn JH, Rehm M. Proteasome inhibition can induce an autophagy-dependent apical activation of caspase-8. Cell Death Differ. 2011;18(10):1584–1597. doi: 10.1038/cdd.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, Lee Y, Park YK, Bae KH, Cho S, Lee dH, Park BC, Kang S, Park SG. HS 1-associated protein X-1 is cleaved by caspase-3 during apoptosis. Mol Cells. 2008;25(1):86–90. [PubMed] [Google Scholar]
- Li B, Hu Q, Xu R, Ren H, Fei E, Chen D, Wang G. Hax-1 is rapidly degraded by the proteasome dependent on its PEST sequence. BMC Cell Biol. 2012;13:20. doi: 10.1186/1471-2121-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M, Nicholson DW, Roy S, Thornberry NA, Peterson EP, Casciola-Rosen LA, Rosen A. The caspase-3 precursor has a cytosolic and mitochondrial distribution: implications for apoptotic signaling. J Cell Biol. 1998;140(6):1485–1495. doi: 10.1083/jcb.140.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser J, Roonprapunt C, Margolis B. C. elegans cell migration gene mig-10 shares similarities with a family of SH2 domain proteins and acts cell nonautonomously in excretory canal development. Dev Biol. 1997;184(1):150–164. doi: 10.1006/dbio.1997.8516. [DOI] [PubMed] [Google Scholar]
- Margolis B, Silvennoinen O, Comoglio F, Roonprapunt C, Skolnik E, Ullrich A, Schlessinger J. High-efficiency expression/cloning of epidermal growth factor-receptor-binding proteins with Src homology 2 domains. Proc Natl Acad Sci USA. 1992;89(19):8894–8898. doi: 10.1073/pnas.89.19.8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda G, Nakajima K, Kawaguchi Y, Yamanashi Y, Hirai K. Epstein-Barr virus (EBV) nuclear antigen leader protein (EBNA-LP) forms complexes with a cellular anti-apoptosis protein Bcl-2 or its EBV counterpart BHRF1 through HS1-associated protein X-1. Microbiol Immunol. 2003;47(1):91–99. doi: 10.1111/j.1348-0421.2003.tb02790.x. [DOI] [PubMed] [Google Scholar]
- McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5(4):a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrione A. Grb10 proteins in insulin-like growth factor and insulin receptor signaling (review) IntJ Mol Med. 2000;5(2):151–154. doi: 10.3892/ijmm.5.2.151. [DOI] [PubMed] [Google Scholar]
- Nantel A, Mohammad-Ali K, Sherk J, Posner BI, Thomas DY. Interaction of the Grb10 adapter protein with the Raf1 and MEK1 kinases. J Biol Chem. 1998;273(17):10475–10484. doi: 10.1074/jbc.273.17.10475. [DOI] [PubMed] [Google Scholar]
- Nantel A, Huber M, Thomas DY. Localization of endogenous Grb10 to the mitochondria and its interaction with the mitochondrial-associated Raf-1 pool. J Biol Chem. 1999;274(50):35719–35724. doi: 10.1074/jbc.274.50.35719. [DOI] [PubMed] [Google Scholar]
- Ooi J, Yajnik V, Immanuel D, Gordon M, Moskow JJ, Buchberg AM, Margolis B. The cloning of Grb10 reveals a new family of SH2 domain proteins. Oncogene. 1995;10(8):1621–1630. [PubMed] [Google Scholar]
- Patel MR, Chang YF, Chen IY, Bachmann MH, Yan X, Contag CH, Gambhir SS. Longitudinal, noninvasive imaging of T-cell effector function and proliferation in living subjects. Cancer Res. 2010;70(24):10141–10149. doi: 10.1158/0008-5472.CAN-10-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudyal P, Shrestha S, Madanayake T, Shuster CB, Rohrschneider LR, Rowland A, Lyons BA. Grb7 and Filamin-a associate and are colocalized to cell membrane ruffles upon EGF stimulation. J Mol Recognit. 2013;26(11):532–541. doi: 10.1002/jmr.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pero SC, Shukla GS, Cookson MM, Flemer S, Krag DN. Combination treatment with Grb7 peptide and Doxorubicin orTrastuzumab (Herceptin) results in cooperative cell growth inhibition in breast cancer cells. Br J Cancer. 2007;96(10):1520–1525. doi: 10.1038/sj.bjc.6603732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter CJ, Wilce MC, Mackay JP, Leedman P, Wilce JA. Grb7-SH2 domain dimerisation is affected by a single point mutation. Eur Biophys J. 2005;34(5):454–460. doi: 10.1007/s00249-005-0480-1. [DOI] [PubMed] [Google Scholar]
- Pradip D, Bouzyk M, Dey N, Leyland-Jones B. Dissecting GRB7-mediated signals for proliferation and migration in HER2 overexpressing breast tumor cells: GTP-ase rules. Am J Cancer Res. 2013;3(2):173–195. [PMC free article] [PubMed] [Google Scholar]
- Radhika V, Onesime D, Ha JH, Dhanasekaran N. Galpha13 stimulates cell migration through cortactin-interacting protein Hax-1. J Biol Chem. 2004;279(47):49406–49413. doi: 10.1074/jbc.M408836200. [DOI] [PubMed] [Google Scholar]
- Ramsay AG, Keppler MD, Jazayeri M, Thomas GJ, Parsons M, Violette S, Weinreb P, Hart IR, Marshall JF. HS1-associated protein X-1 regulates carcinoma cell migration and invasion via clathrin-mediated endocytosis of integrin alphavbeta6. Cancer Res. 2007;67(11):5275–5284. doi: 10.1158/0008-5472.CAN-07-0318. [DOI] [PubMed] [Google Scholar]
- Ritter PM, Marti A, Blanc C, Baltzer A, Krajewski S, Reed JC, Jaggi R. Nuclear localization of procaspase-9 and processing by a caspase-3-like activity in mammary epithelial cells. Eur J Cell Biol. 2000;79(5):358–364. doi: 10.1078/S0171-9335(04)70040-0. [DOI] [PubMed] [Google Scholar]
- Ruzzene M, Penzo D, Pinna LA. Protein kinase CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) induces apoptosis and caspase-dependent degradation of haematopoietic lineage cell-specific protein 1 (HS1) in Jurkat cells. Biochem J. 2002;364(Pt 1):41–47. doi: 10.1042/bj3640041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlberg KK, Hongisto V, Edgren H, Mäkelä R, Hellström K, Due EU, Moen Vollan HK, Sahlberg N, Wolf M, Børresen-Dale AL, Perälä M, Kallioniemi O. The HER2 amplicon includes several genes required for the growth and survival of HER2 positive breast cancer cells. Mol Oncol. 2013;7(3):392–401. doi: 10.1016/j.molonc.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TV, Wang HW, Koumi A, Hollyman D, Endo Y, Ye H, Du MQ, Boshoff C. K15 protein of Kaposi’s sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J Virol. 2002;76(2):802–816. doi: 10.1128/JVI.76.2.802-816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TL, Guan JL. Grb7 in intracellular signaling and its role in cell regulation. Front Biosci. 2004;9:192–200. doi: 10.2741/1229. [DOI] [PubMed] [Google Scholar]
- Shen TL, Han DC, Guan JL. Association of Grb7 with phosphoinositides and its role in the regulation of cell migration. J Biol Chem. 2002;277(32):29069–29077. doi: 10.1074/jbc.M203085200. [DOI] [PubMed] [Google Scholar]
- Siamakpour-Reihani S, Peterson TA, Bradford AM, Argiros HJ, Haas LL, Lor SN, Haulsee ZM, Spuches AM, Johnson DL, Rohrschneider LR, Shuster CB, Lyons BA. Grb7 binds to Hax-1 and undergoes an intramolecular domain association that offers a model for Grb7 regulation. J Mol Recognit. 2011;24(2):314–321. doi: 10.1002/jmr.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siamakpour-Reihani S, Argiros HJ, Wilmeth LJ, Haas LL, Peterson TA, Johnson DL, Shuster CB, Lyons BA. The cell migration protein Grb7 associates with transcriptional regulator FHL2 in a Grb7 phosphorylation-dependent manner. J Mol Recognit. 2009;22(1):9–17. doi: 10.1002/jmr.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen T. Hax-1: a regulator of calcium signaling and apoptosis progression with multiple roles in human disease. Expert Opin Ther Targets. 2011;15(6):741–751. doi: 10.1517/14728222.2011.561787. [DOI] [PubMed] [Google Scholar]
- Slee EA, Adrain C, Martin SJ. Executioner caspase-3, −6, and −7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276(10):7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- Stein D, Wu J, Fuqua SA, Roonprapunt C, Yajnik V, D’Eustachio P, Moskow JJ, Buchberg AM, Osborne CK, Margolis B. The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBOJ. 1994;13(6):1331–1340. doi: 10.1002/j.1460-2075.1994.tb06386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, Lorenzo HK, Zamzami N, Marzo I, Brenner C, Larochette N, Prévost MC, Alzari PM, Kroemer G. Mitochondrial release of caspase-2 and −9 during the apoptotic process. J Exp Med. 1999;189(2):381–394. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Demoliere C, Kitamura D, Takeshita H, Deuschle U, Watanabe T. HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J Immunol. 1997;158(6):2736–2744. [PubMed] [Google Scholar]
- Tanaka S, Mori M, Akiyoshi T, Tanaka Y, Mafune K, Wands JR, Sugimachi K. A novel variant of human Grb7 is associated with invasive esophageal carcinoma. J Clin Invest. 1998;102(4):821–827. doi: 10.1172/JCI2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebinska A, Rembiszewska A, Ciosek K, Ptaszynski K, Rowinski S, Kupryjanczyk J, Siedlecki JA, Grzybowska EA. HAX-1 overexpression, splicing and cellular localization in tumors. BMC Cancer. 2010;10:76. doi: 10.1186/1471-2407-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Lin YL, Tsui YC, Wei LN. Dual action of epidermal growth factor: extracellular signal-stimulated nuclear-cytoplasmic export and coordinated translation of selected messenger RNA. J Cell Biol. 2010;188(3):325–333. doi: 10.1083/jcb.200910083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafiadaki E, Sanoudou D, Arvanitis DA, Catino DH, Kranias EG, Kontrogianni-Konstantopoulos A. Phospholamban interacts with HAX-1, a mitochondrial protein with anti-apoptotic function. J Mol Biol. 2007;367(1):65–79. doi: 10.1016/j.jmb.2006.10.057. [DOI] [PubMed] [Google Scholar]
- Vafiadaki E, Arvanitis DA, Pagakis SN, Papalouka V, Sanoudou D, Kontrogianni-Konstantopoulos A, Kranias EG. The anti-apoptotic protein HAX-1 interacts with SERCA2 and regulates its protein levels to promote cell survival. Mol Biol Cell. 2009;20(1):306–318. doi: 10.1091/mbc.E08-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HG, Rapp UR, Reed JC. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell. 1996;87(4):629–638. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- Yap SV, Vafiadaki E, Strong J, Kontrogianni-Konstantopoulos A. HAX-1: a multifaceted antiapoptotic protein localizing in the mitochondria and the sarcoplasmic reticulum of striated muscle cells. J Mol Cell Cardiol. 2010;48(6):1266–1279. doi: 10.1016/j.yjmcc.2009.10.028. [DOI] [PubMed] [Google Scholar]
- Yokote K, Margolis B, Heldin CH, Claesson-Welsh L. Grb7 is a downstream signaling component of platelet-derived growth factor alpha- and beta-receptors. J Biol Chem. 1996;271(48):30942–30949. doi: 10.1074/jbc.271.48.30942. [DOI] [PubMed] [Google Scholar]
- Zayat V, Balcerak A, Korczynski J, Trebinska A, Wysocki J, Sarnowska E, Chmielarczyk M, Macech E, Konopiński R, Dziembowska M, Grzybowska EA. HAX-1: a novel p-body protein. DNA Cell Biol. 2015;34(1):43–54. doi: 10.1089/dna.2014.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Snipas S, Orth K, Muzio M, Dixit VM, Salvesen GS. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J Biol Chem. 1997;272(12):7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]
- Zorn JA, Wolan DW, Agard NJ, Wells JA. Fibrils colocalize caspase-3 with procaspase-3 to foster maturation. J Biol Chem. 2012;287(40):33781–33795. doi: 10.1074/jbc.M112.386128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.