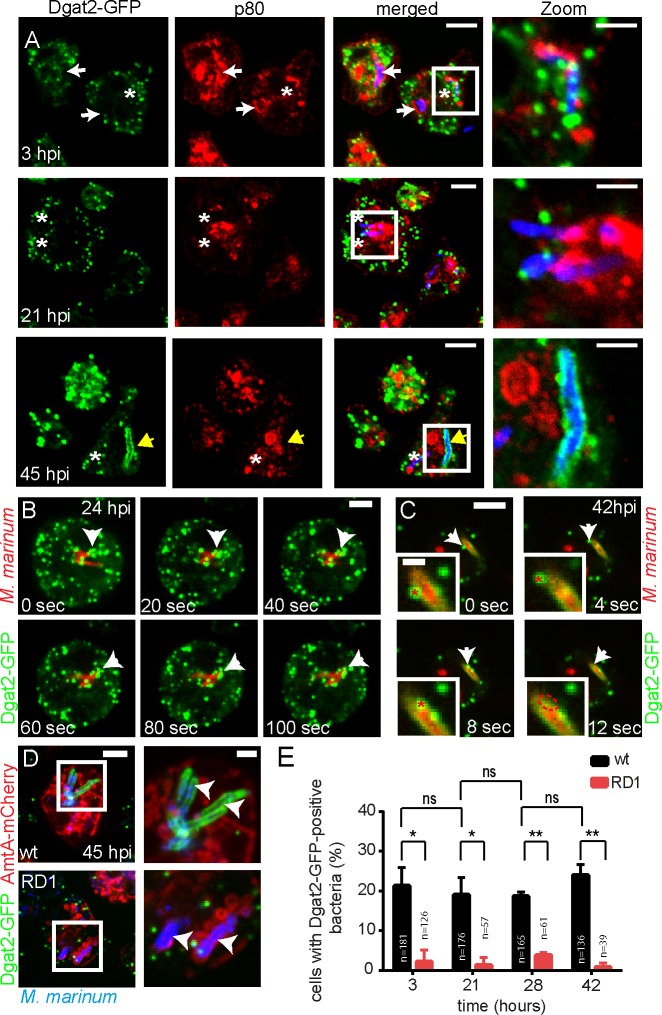
Fig 2. Dynamics of Dgat2-GFP-LDs during infection.
A. Dgat2-GFP-positive LDs attach to the bacteria when they are exposed to the cytosol (3 and 21 hpi). At a later infection stage, Dgat2-GFP completely surrounds a cytosolic bacterium (45 hpi). White arrows point to bacteria that are inside the p80-positive MCV. Asterisks label bacteria that are partially exposed to the cytosol. The yellow arrow points to a cytosolic bacterium that is completely surrounded by Dgat2-GFP. Scale bars, 10 μm; Zoom, 2 μm. B. Dgat2-GFP-labelled LDs stick to an intracellular mCherry-expressing mycobacterium. A time-lapse movie was recorded at 24 hpi with 5 sec frame intervals. Arrows point to LDs aggregated at the surface of the bacterium. Scale bar, 3 μm. C. Coalescence of a Dgat2-GFP-positive LD with an mCherry-expressing mycobacterium surrounded by Dgat2-GFP (arrows). A time-lapse movie was recorded at 42 hpi with 2 sec frame intervals. Arrows point to an LD that coalescences onto the bacterium. Asterisks label the same LD in the Zoom. Scale bar, 5 μm; Zoom, 2μm. D. Dgat2-GFP surrounds a cytosolic wild type M. marinum negative for AmtA-mCherry. No co-localization was observed with M. marinum ΔRD1. Arrows label intracellular bacteria. Samples were taken at 45 hpi and bacteria stained with Vybrant Ruby. Scale bars, 5 μm. E. Quantification of D. While clusters of Dgat2-GFP-labelled LDs were frequently observed close to wild type M. marinum, only a few LDs associated with the RD1 mutant. Dgat2-GFP-positive bacteria were counted in maximum z-projections. Statistical significance was calculated with an unpaired t-test (* p<0.05, ** p<0.01). Bars represent the mean and SD of two independent experiments. For all the experiments presented in Fig 2 Dictyostelium was fed with FA prior to infection.