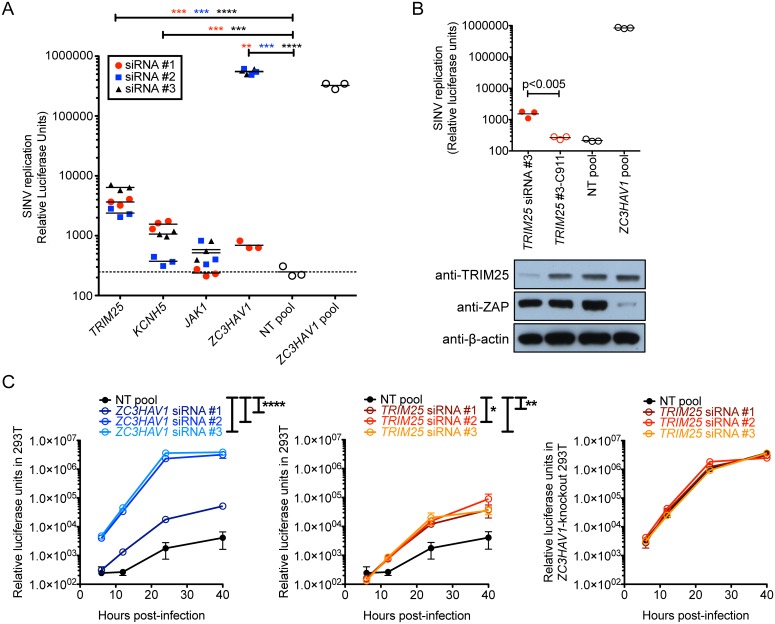
Fig 3. TRIM25 synergizes with ZAP to block SINV replication.
(A) Candidate genes that significantly increased SINV replication in the secondary screen when silenced by individual siRNAs at both concentrations were validated in a larger scale 24-well plate format. Triplicate wells of T-REx-hZAP cells were transfected with the indicated siRNA, induced to express ZAPS, and infected with SINV Toto1101/Luc at a MOI of 10. Each symbol represents the value obtained from a single well after 24 h of infection. White circles represent results using pooled siRNA controls that were either NT or ZC3HAV1-specific. The data is representative of 3 independent experiments. Asterisks indicate statistically significant differences (Student’s t-test, **, p<0.005; ***, p<0.0005; ****, p<0.0001). (B) Triplicate wells of T-REx-hZAP cells were transfected with the indicated siRNA, induced to express ZAPS, and infected with SINV Toto1101/Luc at a MOI of 10. Protein expression levels of TRIM25 and ZAP for the same transfections in a duplicate well were determined by immunoblotting. β-actin was used as a loading control. The data is representative of 4 independent experiments. The p-value from Student’s t-test is shown. (C) SINV replication in infected 293T cells in which ZC3HAV1 (left) or TRIM25 (middle) were silenced, and in ZC3HAV1-null 293T cells in which TRIM25 was silenced (right) is plotted. At 48 h post-transfection with siRNA, cells were infected with SINV Toto1101/Luc at a MOI of 0.01, and lysed at 6, 12, 24, and 40 h p.i. for measurement of luciferase activity. The data is representative of 3 independent experiments. Asterisks indicate statistically significant differences (two-way ANOVA, *, p<0.05; **, p<0.01; ****, p<0.0001).