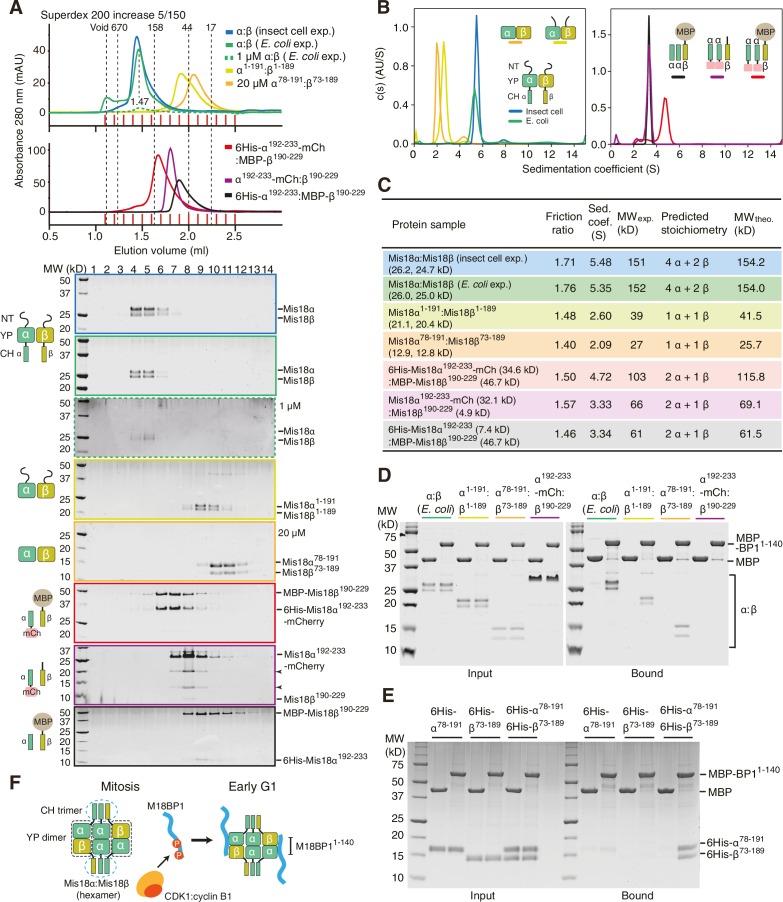
Figure 6. Assembly mechanism of Mis18α:Mis18β-hexamer.
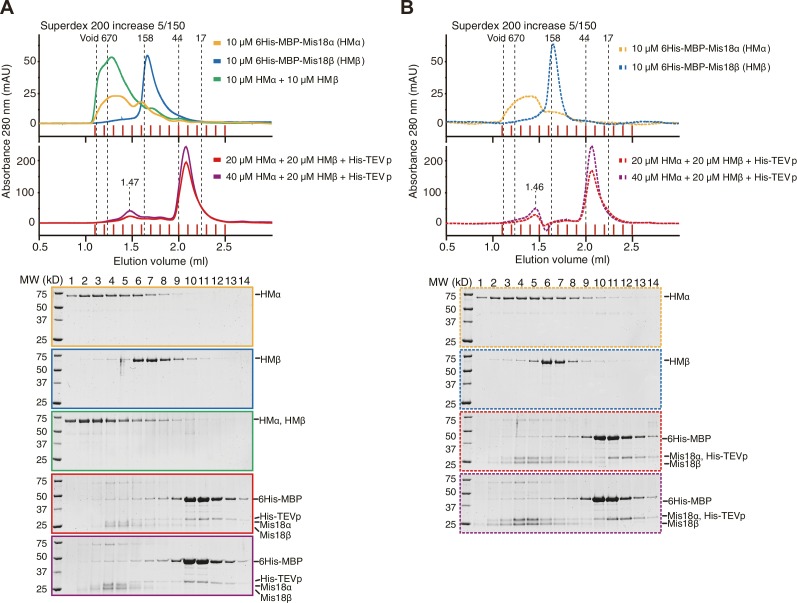
(A) Analytical SEC results of insect-cell-expressed Mis18α:Mis18β (blue), E. coli-expressed Mis18α:Mis18β (green), Mis18α1-191:Mis18β1-189 (yellow-green), Mis18α78-191:Mis18β73-189 (orange), 6His-Mis18α192-233-mCherry:Mis18β190-229 (red), Mis18α192-233-mCherry:Mis18β190-229 (purple), 6His-Mis18α192-233:Mis18β190-229-MBP (black). SEC experiments were carried out with 10 µM protein (loading concentration), unless indicated in the figure. The elution volumes of thyroglobulin (670 kD), aldolase (158 kD), ovalbumin (44 kD), and myoglobin (17 kD) are shown as standards. Red lines indicate fractions collected for Tricine–SDS-PAGE analyses. Gels were stained with CBB, except the gel for 1 µM Mis18α:Mis18β (dashed green line), which was stained with SYPRO Ruby. Left-pointing arrowheads indicate degradation products of Mis18α192-233-mCherry. NT, N-terminal tail; YP, Yippee domain; CH, C-terminal helix; mCh, mCherry. (B) Sedimentation velocity AUC results of the same samples used in the analytical SEC experiments (panel A). The best-fit size distributions are shown with the colors indicated in panel A. Data profiles used for curve-fitting analyses are shown in Figure 6—figure supplement 1. (C) Summary table of the results obtained from the AUC experiments of panel B. Sed. coef., sedimentation coefficient; MWobs., observed molecular weight; MWtheo., theoretical molecular weight. (D, E) Amylose-resin pull-down assays to examine the interaction of Mis18α:Mis18β variants with M18BP11-140. Incubation of amylose beads and proteins (at 5 µM concentration) were performed using a binding buffer containing 30 mM HEPES pH 7.5, 100 mM NaCl, and 1 mM TCEP. Gels were stained with CBB. (F) Hypothetical assembly mechanism of the Mis18α:Mis18β-hexamer in mitosis and the octameric Mis18 complex in the early G1 phase.