Extended Data Figure 10. Reducing mGluRA expression in pacemaker neurons reduces the inhibitory effect of glutamate as well as the siesta.
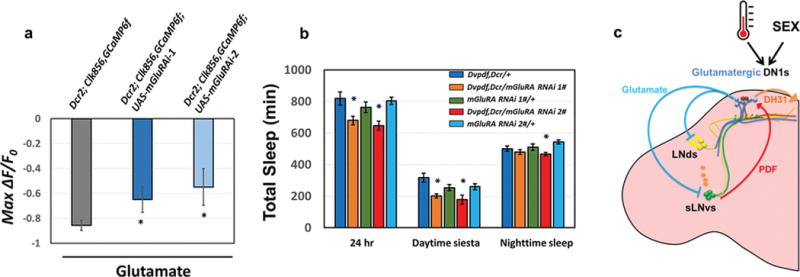
a, The peak decrease of GCaMP6f in circadian cells after applying glutamate to control and mGluRA knockdown flies. The genotypes are shown above the bars. n=6 for UAS-Dcr2; Clk856-GAL4, UAS-GCaMP6f and n=8–9 for UAS-Dcr2; Clk856-GAL4, UAS-GCaMP6f; UAS-mGluRA RNAi groups. ‘*’ indicates p<0.05 by unpaired t-test. Error bars represent SEM. b, Comparison of total sleep, daytime siesta and nighttime sleep in different genotypes. n=32 for each group. ‘*’ indicates p<0.05 by one-way ANOVA with Tukey’s post-hoc test. Error bars represent SEM. c, A temporally constrained negative feedback core pacemaker-DN1 circuit regulates the fly activity/sleep pattern. Early in the day, M pacemaker neurons activate the DN1s via the PDF neuropeptide, and DN1s release DH31 to enhance morning arousal. Later in the day, glutamate release from DN1s inhibits M cells and E cells, promotes the siesta, decreases the evening activity peak and initiates nighttime sleep. A cycling mRNA that encodes inhibitory glutamate receptors in pacemaker cells may help direct this inhibition to the late day. This feedback circadian circuit shapes the bimodal locomotor activity peak and sleep/wake cycles under normal conditions. The higher daily neuronal activity in male DN1s compared to female DN1s promotes the sexually dimorphic activity/sleep pattern. DN1s also integrate environmental information such as temperature to promote sleep plasticity.
