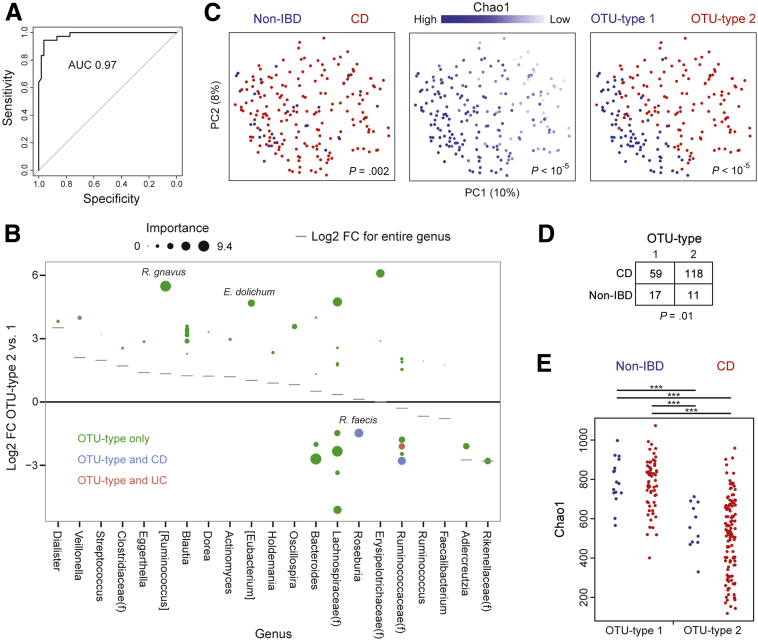
Figure 5.
Microbial community types were predicted in an independent cohort of pediatric CD patients using a random forest classifier. (A) A classifier was created to predict OTU type in the family cohort using 57 OTUs that were differentially abundant in DESeq2 models and were present in at least 30% of samples. The receiver operating characteristic curve of this classifier based on 10-fold cross-validation is shown. (B) Log2 FC between OTU types 2 and 1 in the family cohort for OTUs included in the random forest classifier. Size is proportional to the importance score of the OTU in the classifier, which measures the loss in accuracy of the classifier when the OTU is permutated randomly. (C) PCoA plots visualizing microbial composition in the RISK cohort of pediatric CD patients and controls with gastrointestinal symptoms but no evidence of inflammatory disease. Samples are colored by IBD status, Chao1, or OTU type predicted from the classifier shown in panel A. P values were calculated using Adonis. (D) Contingency table of IBD status by predicted OTU type in the RISK cohort. P value was calculated using the Fisher exact test. (E) Chao1 was determined by IBD status and predicted OTU type in the RISK cohort. P values were calculated using the Mann–Whitney U test. ***P < .0001.