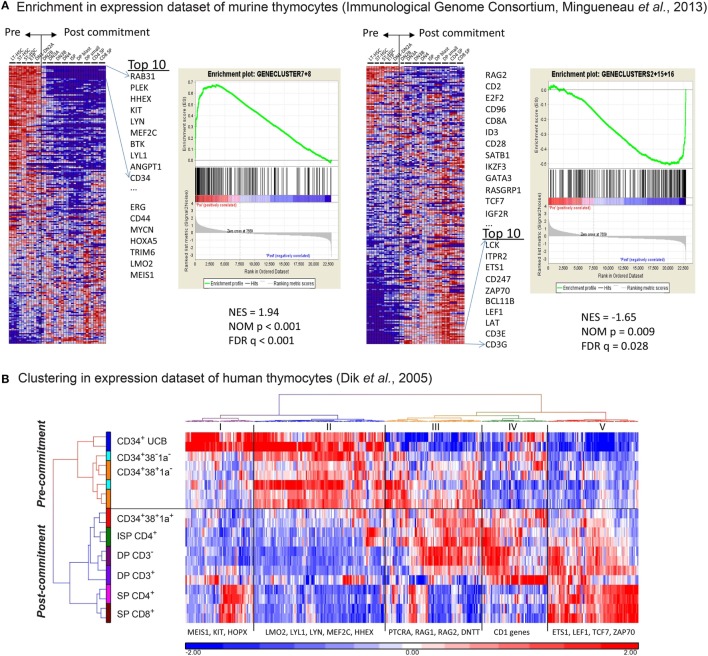
Figure 2.
The in vitro gene expression signatures recapitulate in vivo signatures of pre- and post-T-cell committed thymocytes. (A) Gene set enrichment analysis of our in vitro gene signature on the gene expression dataset of consecutive T-cell development populations from murine thymi (see Table S2 in Supplementary Material). The first and second sub-figures from the left are the analysis results using gene signatures #7 and #8. The Normalized Enrichment Score (NES), Nominal p-value, and false discovery rate q-value indicate that these two gene signatures correlate with the pre-commitment stages. Also shown in the first sub-figure is the list of top 10 input genes with the highest signal-to-noise ratios in this dataset. The third and fourth sub-figures are the same analysis for gene signatures #2, #15, and #16, which correlate with the post-commitment stages. (B) Hierarchical clustering (Euclidean distance and Ward’s method) on human thymocyte populations was performed using 399 out of the 547 in vitro pre-/post-commitment signature genes (see Table S3 in Supplementary Material); the remaining 148 genes were not profiled because they were not on the HG-U133A array used in the dataset in Ref. (16). The dendrogram on the left shows the clustering of different sorted human thymocytes, separating the pre- and post-committed populations.