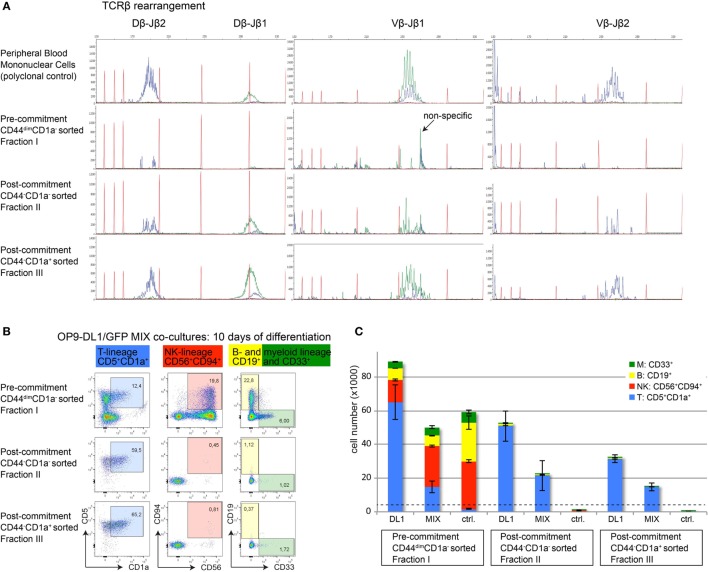
Figure 6.
Loss of CD44 functionally marks human T-cell commitment. (A) GeneScan visualization of in-frame Dβ–Jβ (left column) and Vβ–Jβ (middle and right columns) gene rearrangements, determined using the BIOMED-2 multiplex PCR (19). One representative example of three independent donors is shown. Healthy peripheral blood mononuclear cells were used as a positive, polyclonal control (top row); populations I–III are indicated (rows 2–4). Primers for the Jβ1 cluster were hexachloro-6-carboxy-fluorescein-labeled (blue traces), and primers for the Jβ2 cluster were 6-carboxy-fluorescein (FAM)-labeled (green traces). Red traces: internal size markers. (B) Flow cytometry analysis of sorted human thymocyte populations I–III after 10 days of coculture on OP9-DL1:GFP mixed cells. Differentiation into various hematopoietic lineages is defined as follows: T-lineage (blue): CD5+CD1a+, NK-lineage (red): CD56+CD94+, B-lineage (yellow): CD19+, myeloid lineage (green): CD33+. During differentiation, all populations remain CD45+ and CD7+. (C) Absolute cell numbers ± SD of each differentiated and proliferated hematopoietic lineage from a representative of three experiments, after 10 days of coculture on a layer of OP9-DL1, a 1:1 mix, or OP9-GFP (ctrl.) cells. The number of cells from populations I–III used to initiate the cocultures on day 0 was 4,000 cells/well, indicated by the dashed horizontal line.