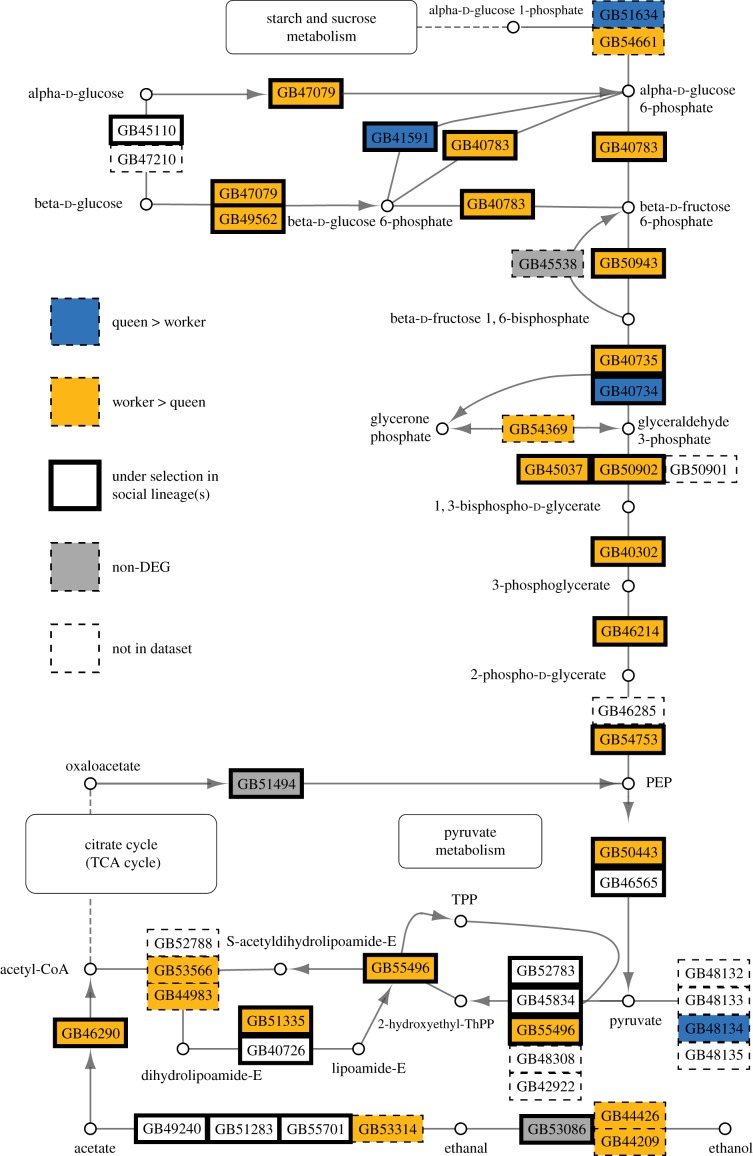
Figure 4.
Differentially expressed genes (FDR < 0.05) between queen and worker M. genalis abdominal tissues in the glycolysis/gluconeogenesis pathway. DEGs were mapped onto putative honeybee orthologues modified from KEGG pathway ame00010 (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/) and [18]. Genes labelled as under selection were identified as undergoing positive selection in at least one of three studies of selection in bees [18–20]. Genes not in dataset are honeybee genes without a BLAST reciprocal best hit to the M. genalis transcriptome or genes that are not expressed in the abdomen of M. genalis.