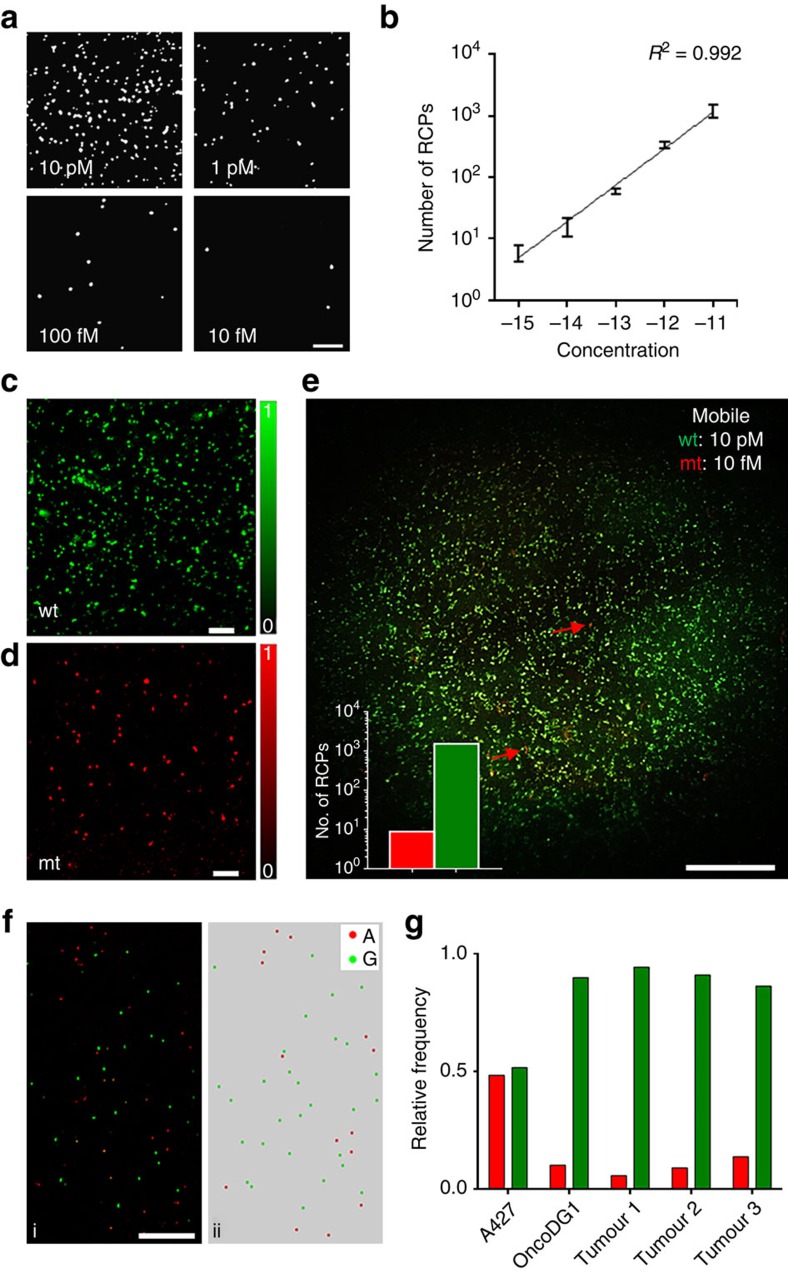
Figure 2. Mobile phone microscopy-based targeted DNA sequencing.
(a) Amplified single-molecule detection through RCA and mobile phone microscopy: selected regions within images of 10 pM to 10 fM RCPs are depicted. Scale bar, 50 μm. (b) Quantification of RCPs generated from a log10 dilution series of synthetic circular templates. Error bars: 1 s.d. from the mean, n=3; linear regression is plotted as straight line. (c) KRAS wild type and (d) KRAS mutant (codon 12 mutation) synthetic DNA fragments were circularized through selector probes, amplified through RCA, and the first base in codon 12 sequenced by unchained SBL chemistry. Sequencing reactions were imaged at both of the fluorescent channels using the mobile phone microscope and these channels were digitally superimposed. Scale bar, 20 μm. (e) Synthetic KRAS fragments at a ratio of 1:1,000 mutant/wild type were sequenced and the reaction imaged through mobile phone microscopy. Cy3 stained RCPs (wild type—base G, green bar) and Cy5 stained RCPs (mutant—base A, red bar) were quantified and plotted in the inset graph. Red arrows in the mobile phone image point to RCPs that show a Cy5 sequencing signal. Scale bar, 200 μm. (f) Mobile phone microscopy-based sequence analysis: (i) fluorescence imaging of sequencing reaction, and (ii) base calling through a custom-written automated image analysis algorithm. Scale bar, 50 μm. (g) Quantification of sequencing experiments of extracted genomic DNA from cell lines and colon tumour biopsies using our mobile phone microscope. Relative frequencies of mutant (red bars) and wild type (green bars) RCPs are plotted.