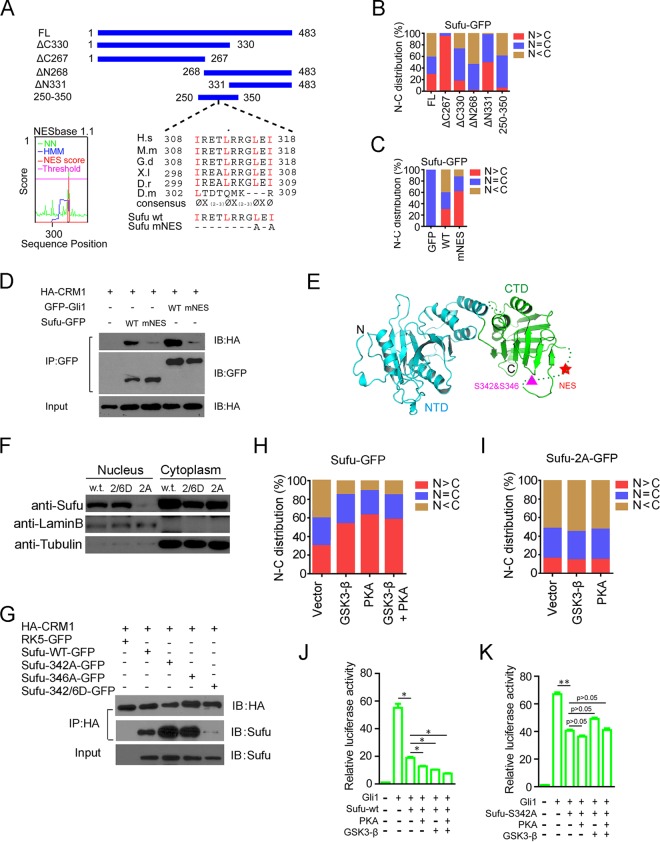
FIG 4.
Identification of a Crm1-mediated nuclear export signal of Sufu and its regulation at a juxtaposing dual phosphorylation site. (A) Schematic representation of amino acid sequences of full-length and deletion mutant Sufu. Alignments of sequences between residues 308 and 318 (mouse) are shown. The mutant mouse Sufu-mNES contains replacements of L316 and I318 by alanine. NESbase 1.1 search scores are shown on the left. (B and C) The nuclear and cytoplasmic distribution (percentage) of various GFP-Sufu deletion constructs (B) and the NES mutant (C) was determined by transient expression in normal MEFs (n ≥ 57). (D) Co-IP analyses of Sufu-GFP and HA-Crm1 interaction in HEK293 cells. GFP-Gli1 was used as a positive control. (E) Three-dimensional structural model of full-length Sufu showing the position of NES and the dual phosphorylation site in the amorphous loop in the C terminus (61). (F) Western analyses of the nuclear and cytoplasmic fractionated samples from transiently transfected cells. Lamin B and tubulin were used as the nuclear and cytoplasmic markers, respectively. (G) Co-IP analyses of interaction between HA-Crm1 and various phosphorylation site mutants of Sufu-GFP. (H and I) Effects of PKA and GSK3β on nucleocytoplasmic distribution of wild-type (H) and the nonphosphorylatable mutant Sufu342A (I) were determined in normal MEFs (n ≥ 65). (J and K) Shh-responsive reporter luciferase assays for the inhibition of Gli1-mediated transcription by wild-type Sufu (J) and the nonphosphorylatable mutant Sufu342A (K).