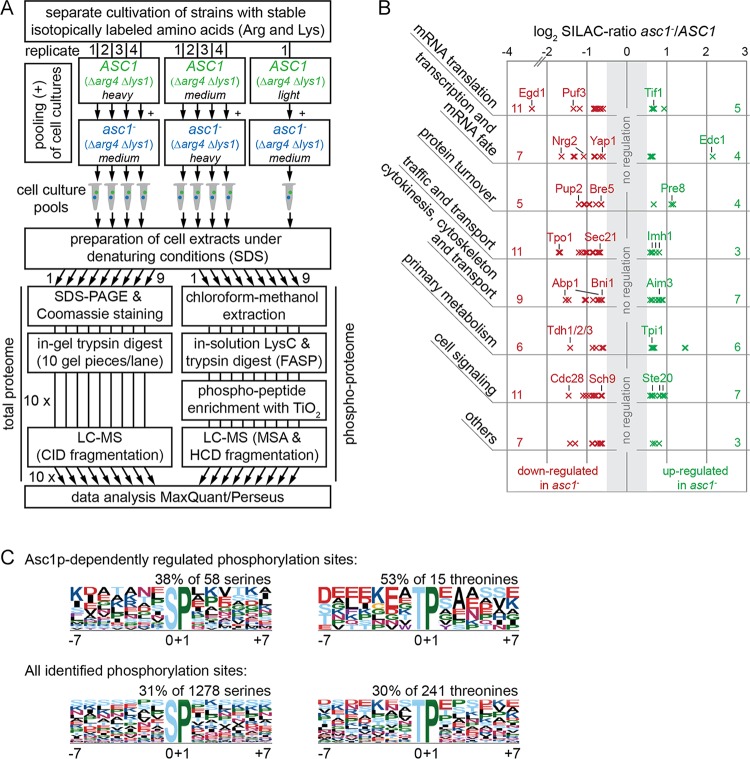
FIG 6.
Analysis of the Asc1p-dependent phosphoproteome. (A) Peptide sample preparation for phosphoproteome and proteome analyses. S. cerevisiae ASC1 wild-type and asc1− strains auxotrophic for arginine and lysine (Δarg4 Δlys1) were cultivated in the presence of lightly, intermediately (medium), or heavily labeled arginine and lysine as indicated in the figure. Arrows indicate which cultures were pooled and show the number of replicates. In total, nine independent cell pools were obtained and subjected to the subsequent preparation of cell extracts. The protein extracts were split in two and processed separately for proteome and phosphoproteome analyses as described in detail in Materials and Methods. For an extended version of this workflow that comprises additional strains, see Fig. S3 in the supplemental material. (B) Assignment of the Asc1p-dependently regulated phosphoproteins to cellular processes. The 90 proteins were grouped in a nonexclusive manner, meaning that 15 proteins are present in two groups. The numbers of proteins assigned to each group are given on the left and right sides of the graph (see Table S7 for the identities of all proteins in the groups). Each cross represents a single regulated phosphosite and indicates its degree of regulation. (C) Overrepresented motifs for phosphorylated serine and threonine residues. Abundances of motifs for Asc1p-dependently regulated phosphosites were compared to the occurrences of the same motifs for all phosphosites identified in this study. Motifs were searched for sites with localization probabilities of ≥0.75 by using motif-x software (49, 50). Additional motifs identified for the complete proteome are not depicted.