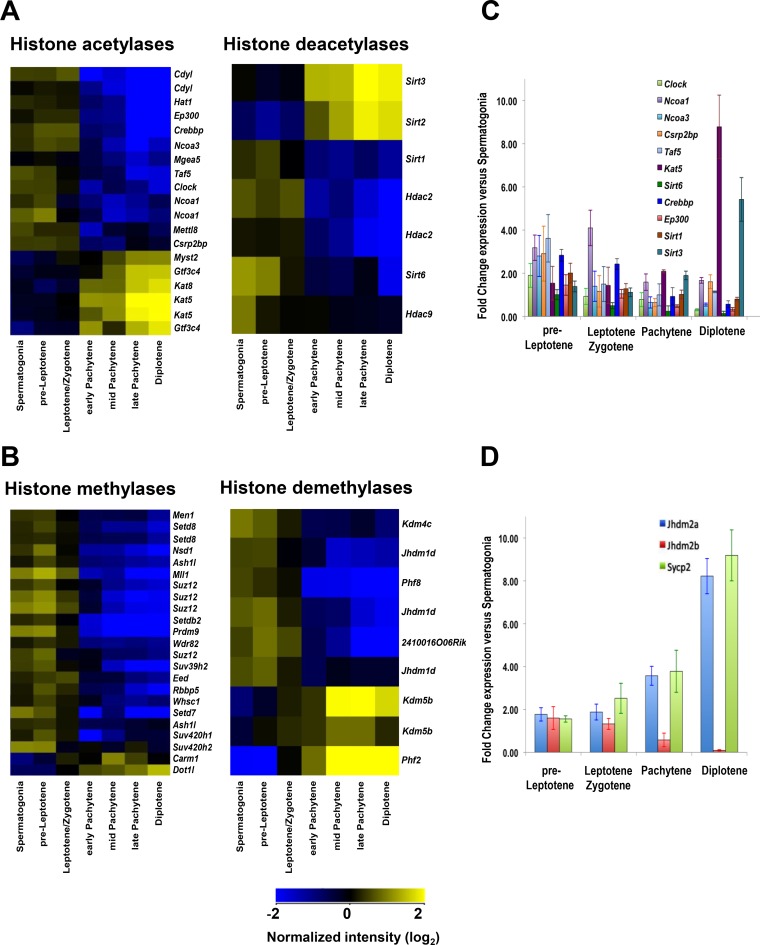
FIG 1.
Expression of histone-modifying enzymes during meiosis. (A) Profiling of differentially expressed genes encoding histone acetylases and deacetylases in all meiosis I cell stages from wild-type B6/DBA mouse testes. (B) Profiling of differentially expressed genes encoding histone methylases and demethylases in all meiosis I cell stages from wild-type B6/DBA mouse testes. (A and B) Meiotic cells were isolated using FACS sorting as described in Materials and Methods. For all heat maps, an absolute fold change filter of 1.5-fold for the spermatogonium/average pachytene expression ratio was applied to obtain differentially expressed genes, followed by hierarchical clustering. Only significantly changed probe sets with P values (corrected) of <0.05 are shown. Microarray data were normalized across the median for all samples, and microarray analyses are described in Materials and Methods. Details of statistical analysis are provided in Data Set S1 in the supplemental material, and the findings were confirmed by qRT-PCR. (C) qRT-PCR analyses of genes encoding HATs and HDACs were performed for all stages of meiosis I. (D) qRT-PCR analyses of specific meiotic stage marker genes that are selectively expressed at different stages of meiosis I (90). (C and D) The y axis shows the normalized expression ratio for the indicated meiotic stage versus expression in spermatogonia. Results are the means and standard errors of the means (SEM) for technical replicates (n = 3). qRT-PCR analysis details are described in Materials and Methods. Real-time PCR primer pairs used for panel C are shown in Table S1.