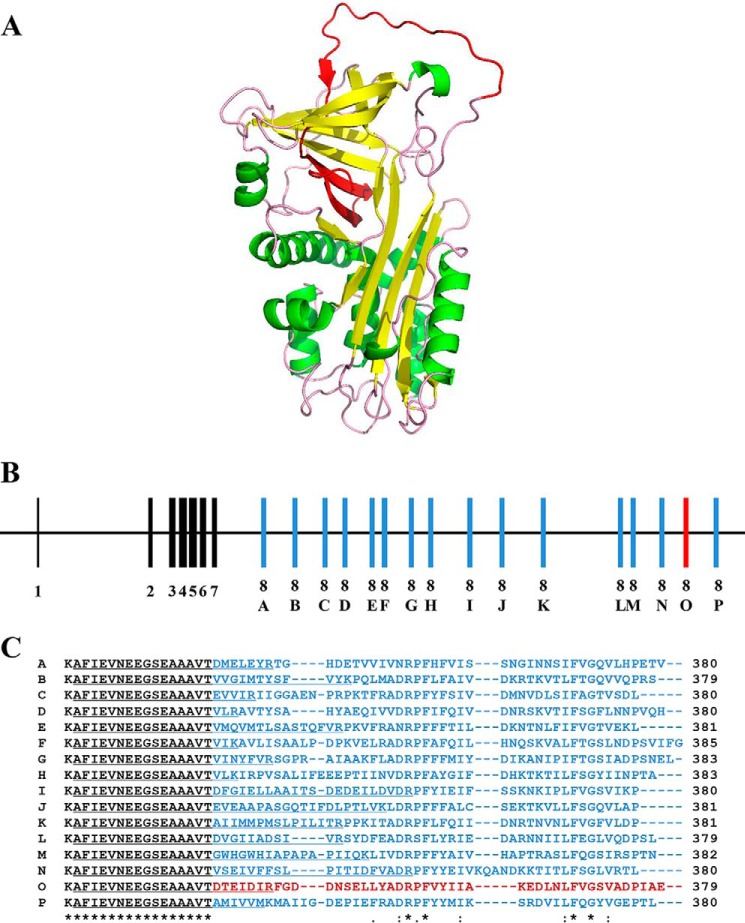
FIGURE 3.
Alternative splicing of the serpin gene for PpS1V with different versions of exon 8 producing different reactive center loops. A, predicted tertiary protein structure of PpS1V. The alternative splicing region in the PpS1V is colored in red. Yellow and green indicate α-helices and β-sheets, respectively. B, structure of the serpin gene for PpS1V. The constant coding regions from exons 1–7 are colored in black. Coding regions of variable exon 8 are colored in red for PpS1V and blue for other isoforms. C, amino acid alignment of the C termini of the 16 predicted isoforms starting with Thr335. The isoform-specific sequences are colored in red for PpS1V and blue for other isoforms. The first isoform-specific tryptic peptides are underlined. An “*” (asterisk) indicates positions that have a single, fully conserved residue. A “:” (colon) indicates conservation between groups of strongly similar properties. A “.” (period) indicates conservation between groups of weakly similar properties.