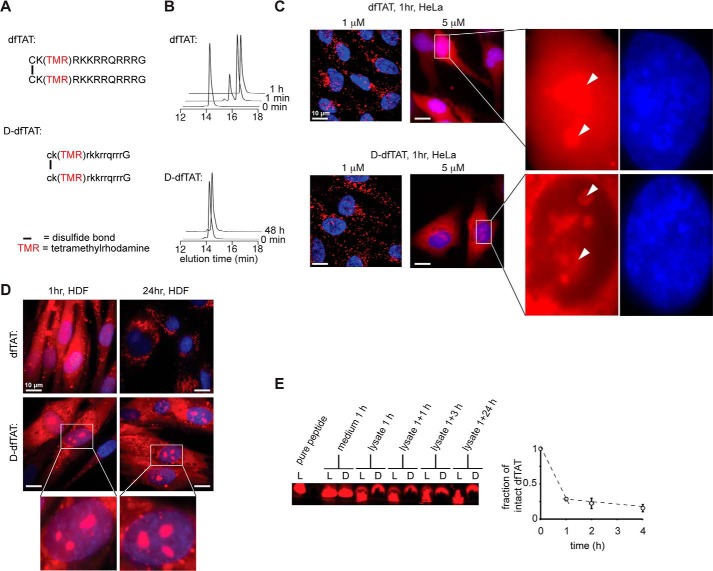
FIGURE 1.
d-dfTAT is protease-resistant, whereas dfTAT is not. A, schematic representation of the amino acid sequence of dfTAT (top) and d-dfTAT (bottom). B, HPLC analysis of dfTAT and d-dfTAT (10 μm) before (0 min time point) and after treatment with trypsin (0.025%). Reactions were quenched at different time intervals with 0.01% TFA in water (1 min, 1 h, and 48 min). Peaks were detected by monitoring the absorbance of the TMR chromophore at 550.8 nm. C, cellular distribution of dfTAT and d-dfTAT after a 1-h incubation. HeLa cells were incubated with dfTAT or d-dfTAT at 1 or 5 μm for 1 h, washed, and then stained with the cell-permeable Hoechst nuclear stain (1 μg/ml). Live cells were imaged with a ×100 objective. Fluorescence images are overlays of the TMR emission at 560 nm (pseudocolored red) and Hoechst emission at 460 nm (pseudocolored blue). Insets to the left show a zoom into the nucleolar staining of each peptide (white arrowheads). Scale bars, 10 μm. D, cellular distribution of dfTAT and d-dfTAT immediately after delivery and after 24 h. HDF cells were incubated with dfTAT or d-dfTAT at 5 μm for 1 h and washed. Cells were imaged with a ×100 objective either immediately after treatment or after an additional 24-h incubation. Fluorescence images are overlays of the TMR emission at 560 nm (pseudocolored red) and Hoechst emission at 460 nm (DNA stain added during imaging, pseudocolored blue). Scale bars, 10 μm. E, analysis of peptide degradation by Tris-Tricine gel electrophoresis of cell lysates. HeLa cells were incubated with 5 μm dfTAT or d-dfTAT for 1 h and lysed either immediately (1 h) or at 1, 3, or 24 h after peptide incubation. Cell lysates were analyzed by electrophoresis along with an aliquot of the peptide solution incubated with cells (medium 1 h) and with a sample of pure peptide. The gel was imaged on a fluorescence scanner. The fluorescent bands were quantified by densitometry to generate the graph presented. The data reported represent the average and corresponding S.D. values (error bars) of biological triplicates.