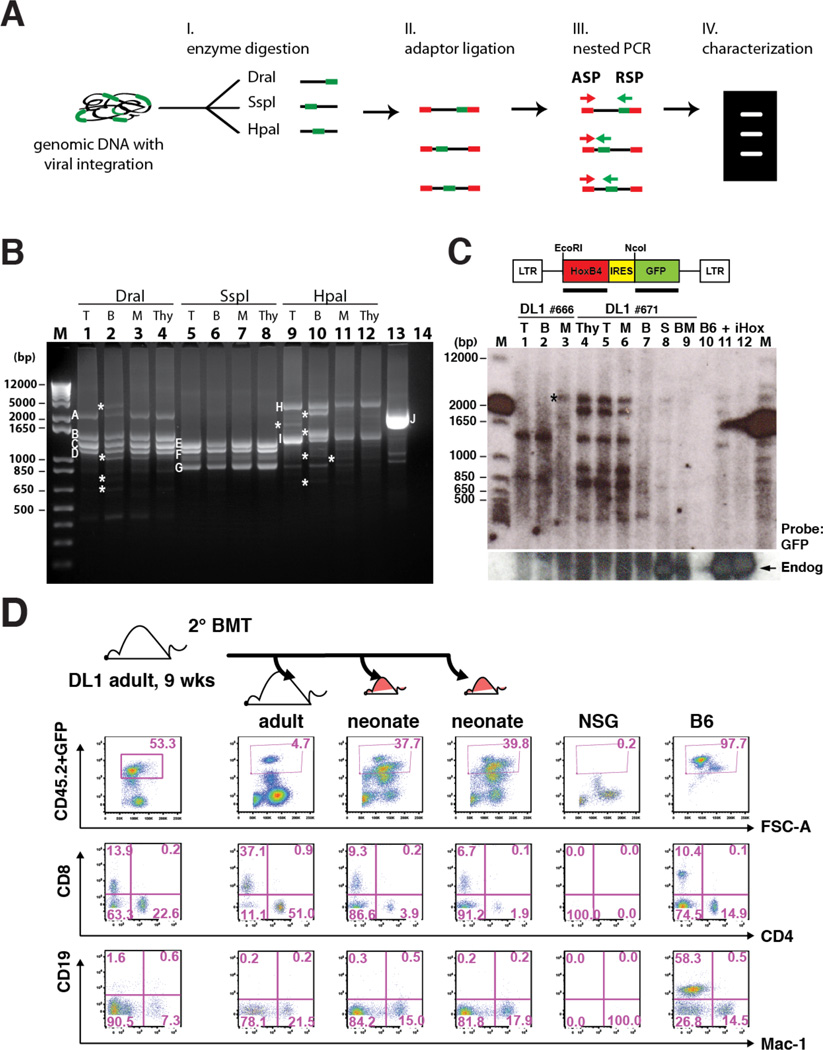
Figure 5. Notch-activated ESC-HSC Clones Serially Reconstitute Multilineage Hematopoiesis.
(A) Schematic diagram of retroviral integration site analysis by LAM-PCR. Genomic DNAs with proviral integrations (green) were digested with three sets of restriction enzyme (DraI, SspI and HpaI) and ligated with adaptors (red). Major amplicons generated by nested PCR with adaptor-specific primer (ASP) and retrovirus-specific primer (RSP) were sequenced and mapped for integration sites. (B) LAM-PCR of splenic donor-derived lineages of a representative iICN ESC-HSC-DL1 recipient at 16 wks post-transplant. Major bands across lineages (A–I) and an assay control of human genomic DNA containing a known retroviral integration at chromosome 17 near RSAD1 gene (J) were individually sequenced (Fig.S5 & Table S1). T: CD3+ T cells; B: B220+ B cells; M: Mac-1+ cells; Thy: thymocytes; #14: no template control; asterisks indicate amplicons predominant in one/two lineages. (C) Southern blotting analysis of common integration sites of two independent recipients (#666 and #671) engrafted with iICN ESC-HSC-DL1. S: splenocytes; BM: bone marrows; B6: B6 splenocytes; +: total #671 splenocyte; iHox: inducible HoxB4 ESC. Top: GFP probed blot; bottom: HoxB4 probed blot. Arrow: endogenous HoxB4; asterisk indicates the clone predominantly detected in one lineage. (D) At 9 wks post-transplant, BMs of a representative iICN ESC-HSC-DL1 recipient were serially transplanted into one adult and two neonatal secondary recipients; lineage analysis of donor-derived reconstitution at 8 wks post-secondary transplant is shown. See also Fig.S5–6 & Table S1.