Abstract
Background
During embryogenesis, tight regulation of retinoic acid (RA) availability is fundamental for normal development. In parallel to RA synthesis, a negative feedback loop controlled by RA catabolizing enzymes of the cytochrome P450 subfamily 26 (CYP26) is crucial. In vertebrates, the functions of the three CYP26 enzymes (CYP26A1, CYP26B1, and CYP26C1) have been well characterized. By contrast, outside vertebrates, little is known about CYP26 complements and their biological roles. In an effort to characterize the evolutionary diversification of RA catabolism, we studied the CYP26 genes of the cephalochordate amphioxus (Branchiostoma lanceolatum), a basal chordate with a vertebrate-like genome that has not undergone the massive, large-scale duplications of vertebrates.
Results
In the present study, we found that amphioxus also possess three CYP26 genes (CYP26-1, CYP26-2, and CYP26-3) that are clustered in the genome and originated by lineage-specific duplication. The amphioxus CYP26 cluster thus represents a useful model to assess adaptive evolutionary changes of the RA signaling system following gene duplication. The characterization of amphioxus CYP26 expression, function, and regulation by RA signaling demonstrated that, despite the independent origins of CYP26 duplicates in amphioxus and vertebrates, they convergently assume two main roles during development: RA-dependent patterning and protection against fluctuations of RA levels. Our analysis suggested that in amphioxus RA-dependent patterning is sustained by CYP26-2, while RA homeostasis is mediated by CYP26-1 and CYP26-3. Furthermore, comparisons of the regulatory regions of CYP26 genes of different bilaterian animals indicated that a CYP26-driven negative feedback system was present in the last common ancestor of deuterostomes, but not in that of bilaterians.
Conclusions
Altogether, this work reveals the evolutionary origins of the RA-dependent regulation of CYP26 genes and highlights convergent functions for CYP26 enzymes that originated by independent duplication events, hence establishing a novel selective mechanism for the genomic retention of gene duplicates.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-016-0863-1) contains supplementary material, which is available to authorized users.
Keywords: Branchiostoma lanceolatum, Cephalochordate, Duplication-Degeneration-Complementation (DDC) model, Lancelet, Retinoic acid (RA) signaling
Background
During animal development, the vitamin A-derived morphogen retinoic acid (RA) mediates a number of crucial functions, including, for example, early embryonic patterning and organogenesis, by acting on different cellular processes ranging from proliferation to cell death [1–7]. In vertebrates, normal development requires a very tightly controlled balance of the total amount of available RA, which is maintained through positive and negative feedback loops associated, respectively, with RA production (chiefly by RALDH1, 2, and 3, for retinaldehyde dehydrogenase 1, 2, and 3) and RA degradation (chiefly by CYP26A1, B1, and C1, for cytochrome P450 subfamily 26A1, B1, and C1) [8–12]. The biological response to endogenous RA, in turn, is mediated by heterodimers of two nuclear receptors, the retinoic acid receptor (RAR) and the retinoid X receptor (RXR), with the expression levels of RAR in particular being tightly linked to the availability of RA [1, 3, 4]. RAR/RXR heterodimers directly exert their transcriptional function by binding to RA response elements (RAREs) in the regulatory regions of RA target genes [13]. A typical RARE is composed of two direct repeats (DRs) corresponding to a conserved nucleotide sequence [(A/G)G(G/T)TCA] separated by a spacer composed of one, two or five nucleotides (corresponding to, respectively, DR1, DR2, and DR5 elements) [13–15]. Upon RA binding, RAR/RXR heterodimers generally function as ligand-activated transcription factors, but can also mediate RA-dependent repression of target genes in a context-specific manner, the exact molecular modalities of which still remain to be established [16].
During vertebrate development, the RA degrading enzymes of the CYP26 subfamily play critical roles in the formation of an anterior-posterior (A-P) RA gradient as well as in the compensation of RA level fluctuations by oxidizing RA into biologically inactive compounds [17]. They are thus characterized by dynamic, yet highly specific, developmental expression patterns in vertebrates [18], with CYP26A1, for example, being expressed in the anterior ectoderm in the early embryo and subsequently becoming localized, amongst other tissues, to the hindbrain, the pharyngeal arches, and the tail bud. Similarly, both CYP26B1 and CYP26C1 are detectable in specific rhombomeres of the hindbrain and in pharyngeal arches as well as in fin and limb buds of the developing embryo [18]. Concomitantly, the loss of CYP26 function has been associated both with A-P patterning defects, most prominently in the developing central nervous system (CNS) and the mesoderm, and an increased sensitivity to RA teratogenicity [19]. For instance, CYP26A1 knockout mice are characterized by a posteriorization of the hindbrain and the vertebral column, and CYP26B1 genetic ablation leads to craniofacial and limb malformations [20]. Interestingly, while the loss of CYP26C1 alone does not result in overt anatomical abnormalities [21], the combined removal of CYP26C1 with either CYP26A1 or CYP26B1 induces phenotypes that are more severe than those aforementioned, thereby suggesting that CYP26C1 plays an important cooperative role in the CYP26-mediated control of endogenous RA levels during vertebrate development [19, 21].
In line with this cooperative action of CYP26 enzymes, the vertebrate RA signaling system in general is characterized by complex feedback mechanisms that are mediated, either indirectly or directly, by RAR/RXR-dependent signaling. As an example of an indirect regulation, it has been shown that, in the vertebrate trunk, RA, generated by RALDH activity, represses and confines FGF8 expression to rostral and caudal domains (i.e. to the heart- and tail bud-associated progenitor fields) [16]. This action is mediated by RAR/RXR heterodimers binding to a repressive DR2 RARE located upstream of the FGF8 gene [22, 23] that, in turn, activates CYP26 expression both anteriorly and posteriorly to limit the extent of RA activity [16, 24, 25]. In addition, the expression of CYP26A1 and CYP26B1 have been shown to be dependent on RA activity, thereby generating a CYP26-controlled negative feedback loop in RA sensitive tissues to reduce the overall amount of available RA [9, 18, 26, 27]. For CYP26A1, this regulation is directly mediated by RAR/RXR heterodimers binding to DR5 RAREs in the promoter region, while for CYP26B1 this control seems to be indirect [26, 27]. Note further that the vertebrate CYP26C1 gene is likely to contribute differently than its paralogs to this negative feedback system, as CYP26C1 expression is actually downregulated following RA stimulation [9].
The intricate molecular mechanisms controlling the catabolism of endogenous RA during vertebrate development likely arose at the base of this lineage following the whole genome duplication (WGD) events that took place during early vertebrate diversification [28, 29]. Therefore, the evolutionary elaboration of the RA signaling system in general seems to be tightly linked to the duplication of RA metabolism genes. The so-called DDC model (for Duplication-Degeneration-Complementation) predicts three possible outcomes following duplication of a gene: non-functionalization (i.e. the loss of one of the duplicates), neo-functionalization (i.e. one of the copies retains the ancestral role, while the other duplicate assumes a novel functionality) or sub-functionalization (i.e. both duplicates assume a part of the function of the single ancestral gene) [30, 31]. While the model predicts that the most likely outcome following duplication of a gene is the loss of one of the duplicates (i.e. non-functionalization), very clear examples for the neo-functionalization and the sub-functionalization of duplicated genes remain scarce [32, 33].
In order to develop a credible scenario for the evolutionary diversification of the vertebrate RA system and investigate the implications of the DDC model in the duplication of RA metabolism genes, we decided to study the function and regulation of RA degradation during embryonic development of the cephalochordate amphioxus (Branchiostoma lanceolatum). Due to its phylogenetic position at the base of chordates, amphioxus is a very useful model to characterize chordate- and vertebrate-specific innovations, both on a morphological and a genomic level. For instance, at the morphological level, amphioxus and vertebrates share a dorsal CNS, a postanal tail as well as pharyngeal gill slits [34, 35], while, conversely, amphioxus lacks some vertebrate-specific characters, such as definitive neural crest and placodes as well as a cartilaginous or bony skeleton [34, 35]. Furthermore, amphioxus is a basal chordate that did not undergo WGD [36, 37] and that possesses a vertebrate-like RA signaling pathway [29, 36]. Thus, while RA signaling in vertebrates is generally controlled by three RARs (RARα, RARβ, RARγ) and three RXRs (RXRα, RXRβ, RXRγ) that form a multitude of different heterodimers, the amphioxus genome contains only one RAR and one RXR gene [38]. Nevertheless, administration of exogenous RA during amphioxus gastrulation leads, as observed in vertebrates, to the posteriorization of the amphioxus CNS and endoderm, hence preventing, for example, the formation of mouth and gill slits [38–43]. These regionalization defects are further associated with a deregulation of RAR and Hox gene expression, which have been shown to be direct targets of RA signaling in amphioxus, as they are in vertebrates [44].
In amphioxus, three CYP26 genes (CYP26-1, CYP26-2, and CYP26-3) have been reported, which are clustered together in the genome and have possibly emerged from a lineage-specific duplication [29]. This CYP26 locus offers a rare, if not unique, opportunity to investigate the adaptive changes following lineage-specific duplication that led to the retention of three CYP26 genes in the genome. The results from our analyses thus show that the three amphioxus CYP26 genes arose by lineage-specific tandem duplication of a single, ancestral CYP26 gene. They further provide evidence that these three genes assume two main functions during amphioxus development, as they do in vertebrates, i.e. patterning of the embryo and protection against RA level fluctuations. These two roles have been sub-functionalized in amphioxus with CYP26-2 mediating RA-dependent developmental patterning and CYP26-1 and CYP26-3 assuming the protection of the embryo from RA teratogenesis. Moreover, the presence of functional RAREs in the amphioxus CYP26 cluster indicates that RA degradation is regulated in cephalochordates like in vertebrates, i.e. directly by RAR/RXR heterodimers, hence establishing a negative RA feedback system. Comparative genomic analyses of CYP26 regulatory regions from different bilaterian animals further suggest that this CYP26-dependent negative RA feedback system is not unique to chordates, but probably arose earlier in animal evolution and was already present in the last common ancestor of all deuterostomes, but not in that of all bilaterians. The adaptive advantages of an elaborate CYP26-driven RA degradation system are discussed. In sum, the evolutionary history of amphioxus CYP26 genes provides an excellent example for the sub-functionalization of two distinct developmental functions and a paradigm for understanding the selective mechanisms acting on duplicated genes and leading to their retention in the genome.
Results
CYP26 genes were duplicated independently several times in bilaterian evolution
Previous analyses have reported three CYP26 genes in the Florida amphioxus, Branchiostoma floridae, and have suggested that they likely originated by lineage-specific duplication from a single ancestral CYP26 gene [29, 36]. Here, we have identified and cloned three CYP26 genes from the European amphioxus, Branchiostoma lanceolatum. To further assess the phylogenetic relationships of the amphioxus CYP26 genes relative to each other and to other members of the CYP26 subfamily, thereby distinguishing between orthologous and paralogous CYP26 genes, we first carried out phylogenetic analyses using as outgroup the CYP51 genes, which constitute the CYP subfamily that is most closely related to the CYP26 genes [45]. For this phylogenetic tree reconstruction, we used all CYP26 sequences from 15 vertebrates and 13 invertebrates, including three cephalochordate species (B. lanceolatum, B. floridae, and B. belcheri). Of note, while we successfully identified genes encoding CYP26 in the genomes of priapulids, brachiopods, mollusks, annelids, sea urchins, hemichordates, cephalochordates, ascidian tunicates, and vertebrates, we were unable to do so in those of nematodes and arthropods (as previously reported [2, 46]).
Within vertebrates, we found three CYP26 paralogs in both cyclostomes (CYP26A1, CYP26B1/C1a, and CYP26B1/C1b) and gnathostomes (CYP26A1, CYP26B1, and CYP26C1). Multiple CYP26 paralogs were also identified in most invertebrates species studied (two in Capitella teleta, Ciona intestinalis, Lottia gigantea, Priapulus caudatus, and Saccoglossus kowalevskii, three in B. lanceolatum, B. floridae, B. belcheri, Crassostrea gigas, and Ptychodera flava, and four in Lingula anatina), with the notable exceptions of the cephalopod Octopus bimaculoides and the sea urchin Strongylocentrotus purpuratus, each of which possesses only a single CYP26 gene (Additional file 1).
The results of the phylogenetic analysis (Fig. 1 and Additional file 2), obtained with both the Bayesian Inference (BI) and the Maximum Likelihood (ML) methods, suggested an early phylogenetic separation of the vertebrate CYP26A1 sequences from the vertebrate CYP26B1/C1 sequences. Vertebrate CYP26A1 and CYP26B1/C1 thus formed two independent clades within the CYP26 subfamily, both of which being strongly supported: 0,91/96 (posterior probability/bootstrap percentage) for CYP26A1 and 1/99 for CYP26B1/C1. Within these two vertebrate CYP26 clades, the cyclostome sequences consistently branched at the base: Lethenteron japonicum CYP26A1 at the base of the vertebrate CYP26A1 (0,96/98) and CYP26B1/C1a and CYP26B1/C1b from L. japonicum and Petromyzon marinus at the base of the vertebrate CYP26B1/C1 (0,7/58). Within the vertebrate CYP26B1/C1 clade, the association of the gnathostome CYP26B1 sequences (0,73/58) was less robustly supported than that of the gnathostome CYP26C1 sequences (1/95), which might be related to the presence of chondrichthyan-specific CYP26B1 duplicates (CYP26B1 and CYP26B2 from Callorhinchus milii and Leucoraja erinacea) disrupting the base of the CYP26B1 branch. Of note, while our analysis revealed the general presence of CYP26A1, CYP26B1, and CYP26C1 paralogs in chondrichthyans, we were unable to identify a CYP26A1 gene in C. milii and a CYP26C1 gene in L. erinacea. Altogether, these data suggest that the diversification of vertebrate CYP26 genes was a highly complex process, involving WGD, lineage-specific duplications as well as secondary gene losses.
Fig. 1.
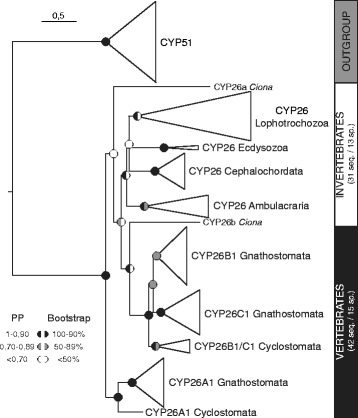
Phylogenetic analysis of the CYP26 subfamily. Diagrammatic summary of Bayesian Inference (BI) and Maximum Likelihood (ML) analyses of the phylogenetic relationships within the CYP26 subfamily, with CYP51 used as outgroup. The detailed tree is shown in Additional file 2 and sequence information is given in Additional file 1. Branch lengths are representative of the amino acid substitution rate and branch support is indicated at each major node as posterior probabilities (PP) for the BI tree and as bootstrap percentages for the ML tree. Furthermore, the total number of sequences (seq.) and species (sp.) is provided for the CYP26 subfamily
Outside vertebrates, the CYP26 sequences from ecdysozoans, ambulacrarians, and cephalochordates always grouped together with very strong support values: the ecdysozoan P. caudatus (1/100), the ambulacrarians P. flava and S. kowalevskii (1/99), and the cephalochordates B. lanceolatum, B. floridae and B. belcheri (1/99). Collectively, these results suggest that, within each of these invertebrate groups, the CYP26 gene complement originated independently by linage-specific duplication. In contrast, the two sequences from the tunicate C. intestinalis did neither associate with each other, nor reliably with one of the major CYP26 clades in the tree. It is therefore impossible to comment on the nature and origin of the CYP26 duplication in this animal. Similarly, the reconstruction of the evolutionary history of lophotrochozoan CYP26 genes is complicated by the lack of phylogenetic resolution between the sequences from the five analyzed lophotrochozoan species, which formed an unresolved polytomy in our analysis. Nonetheless, there is evidence for lineage-specific duplications of CYP26 genes in the brachiopod L. anatina, which possesses four CYP26 paralogs that established two distinct clades in the tree, one very strongly (1/100) and one very weakly (0,8/--) supported. Furthermore, two of the three CYP26 sequences from the oyster C. gigas are grouped within in a single clade, but the support for this association is very weak (0,95/--). Future studies will thus have to address the processes underlying the evolution of lophotrochozoan CYP26 genes.
To gain further insights into the diversification of CYP26 genes in different animal lineages, we next conducted a phylogenetic dating analysis (Additional file 3). This survey indicated that the CYP26 genes of the ecdysozan P. caudatus were likely duplicated independently at the end of the Ordovician (about 446 Mya). Similarly, within the ambulacrarians, we found evidence for linage-specific duplications in hemichordates, which likely occurred during two different periods: the early Carboniferous for S. kowalevskii (about 342 Mya) and the middle Triassic for P. flava (about 240 Mya). Finally, in chordate lineages, CYP26 genes have also likely been duplicated independently in cephalochordates, tunicates, and vertebrates, with the three cephalochordate genes resulting from an initial duplication in the early Carboniferous (about 358 Mya), followed by a subsequent duplication during the middle Permian (about 295 Mya). In contrast, while it is difficult to conclude on the timing of the duplication giving rise to the two CYP26 genes in the ascidian C. intestinalis, the evolution of the vertebrate CYP26 complement is complex and implies a series of duplications, including an ancient split into CYP26A1 and CYP26B1/C1 and the subsequent diversification of CYP26A1, CYP26B1, and CYP26C1 during the Cambrian period (about 548 to 510 Mya). It should be added that, consistent with the results of the phylogenetic tree, the dating analysis did not yield reliable information on the timing of the duplications of lophotrochozoan CYP26 genes.
CYP26-2 expression is suggestive of a function in developmental patterning of the B. lanceolatum embryo
In vertebrates, CYP26A1, CYP26B1, and CYP26C1 have very distinct expression patterns with several key domains being conserved between different species [6, 18]. Thus, we next assessed the temporal and spatial distribution of the three cephalochordate-specific duplicates in the amphioxus B. lanceolatum by in situ hybridization (ISH). The ISH results revealed that the expression profiles of CYP26-1 and CYP26-3 are generally quite similar (Fig. 2a-e, t-x). For both genes, no signal was detectable by ISH from fertilization through mid gastrulation. Expression of both CYP26-1 and CYP26-3 is first identifiable at late gastrula stages as a weak signal in the lateral anterior mesoderm (Fig. 2a, b, t and u). As development proceeds, this domain becomes associated with the most anterior somites at mid neurula stage (Fig. 2c, d, v and w). At this stage, CYP26-1 and CYP26-3 are also discreetly and transiently expressed in the anterior central nervous system (CNS), at the level of the first somite (Fig. 2c and v). Expression of both genes remains very weak and chiefly associated with mesodermal tissues during subsequent developmental stages (Fig. 2e and x): while the CYP26-1 signal is most evident in central and posterior regions of the larva (Fig. 2e), CYP26-3 is mainly detectable in central and more anterior larval territories (Fig. 2x).
Fig. 2.
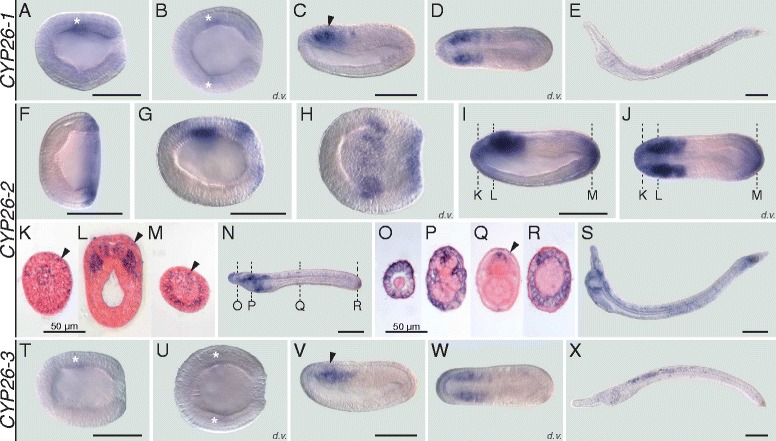
Developmental expression patterns of amphioxus CYP26 genes. Whole mount in situ hybridization experiments were carried out for CYP26-1 (a-e), CYP26-2 (f-s), and CYP26-3 (t-x). Amphioxus (Branchiostoma lanceolatum) embryos and larvae are shown as lateral views with anterior to the left and the dorsal side up, excepting for (b, d, h, j, u, w), which are dorsal views (d.v.) with anterior to the left, and for (k-m) and (o-r), which are cross-sections viewed from the front. Cross-sections in (k-m) are through the embryo shown in (i, j) at the levels indicated by the dashed lines and cross-sections in (o-r) are through the embryo shown in (n) at the levels indicated by the dashed lines. White asterisks in (a, b, t, u) highlight inconspicuous early expression domains of CYP26-1 and CYP26-3. Arrowheads in (c, v) indicate central nervous system expression and in (k-m, q) ectodermal signal. Developmental stages shown are: early gastrula (f), late gastrula (a, b, g, h, t, u), mid neurula (c, d, i-m, v, w), very early larva (n-r), early (60 h) larva (e, s, x). Scale bars are 100 μm for the whole mounts and 50 μm for the cross-sections
In contrast to CYP26-1 and CYP26-3, CYP26-2 has a much more complex developmental expression profile during B. lanceolatum embryonic and early larval development. Expression of CYP26-2 is first detectable by ISH at the mid gastrula stage with the signal being localized globally around the blastopore (Fig. 2f). The blastopore-associated signal subsequently weakens and, at the late gastrula stage, an additional expression domain appears in a region corresponding to presumptive lateral mesoderm and anterior neuroectoderm (Fig. 2g and h). By the mid neurula stage, the mesodermal signal has been expanded into the two anterior-most somite pairs (Fig. 2i, j and l). At this stage, CYP26-2 is further still detectable in the anterior neuroectodem (Fig. 2i, j and l). Additionally, the gene is now expressed in the ectoderm, most conspicuously in the anterior and posterior tips of the embryo (Fig. 2i, j and k-m), as well as in the anterior- and posterior-most endoderm (Fig. 2i, j and m). At the mid neurula stage, the blastopore-associated signal becomes perceivable in the newly formed tail bud (Fig. 2f-j and m). In very early larvae, just before the opening of the larval mouth, expression of CYP26-2 is detectable anteriorly in all germ layers, i.e. the ectoderm, the mesoderm, the endoderm as well as in the CNS, with the signal being least noticeable in the endoderm (Fig. 2n-p). Furthermore, in both ectoderm and CNS, individual cells are labeled along the A-P body axis (Fig. 2n and q) and, at the posterior end of the embryo, the tail ectoderm strongly expresses CYP26-2 (Fig. 2n and r). In the amphioxus larva, the overall domains of CYP26-2 expression are maintained, with conspicuous labeling anteriorly and posteriorly and a weaker signal in the center (Fig. 2s). In sum, while CYP26-1 and CYP26-3 expression is very discreet and chiefly limited to the mesoderm, that of CYP26-2 is detectable in all germ layers and dynamically changes in space and time throughout development.
CYP26 acts as a fine regulator of RA levels in the patterning of the B. lanceolatum larval tail fin
Disruption of CYP26 activity causes very severe defects during vertebrate development [18]. To determine the role of CYP26 enzymes in the amphioxus embryo, we subsequently disrupted endogenous RA degradation during amphioxus development by treatments with the CYP26-specific inhibitor R115866 [47, 48]. For comparisons, the R115866 treatments were carried out in parallel to treatments with RA or with two different RAR antagonists (BMS009 and BMS493). The capacity of R115866 to inhibit endogenous RA degradation was verified by double treatments of R115866 and BMS493. The results obtained from these different pharmacological treatments of B. lanceolatum embryos are in large agreement between each other and with previous studies that characterized the roles of RA signaling in amphioxus endoderm specification and pharyngeal patterning [38–43]. Thus, while the downregulation of RA signaling activity by RAR antagonists (1 μM of either BMS009 or BMS493) resulted in an enlarged pharynx and an expansion of pharyngeal structures (Fig. 3e and f), the upregulation of endogenous RA signaling by 1 μM RA led to a shortening of the pharynx and the malformation of pharyngeal structures (such as the mouth and the gill slits) (Fig. 3b). Consistently, the local upregulation of RA signaling by 0,5 μM R115866 yielded similar results (Fig. 3c), and co-treatments of 0,5 μM R115866 and 1 μM BMS493 led to an attenuation of the severe phenotype induced by 0,5 μM R115866 alone with at least a partial recovery of pharynx formation and patterning (Fig. 3d).
Fig. 3.
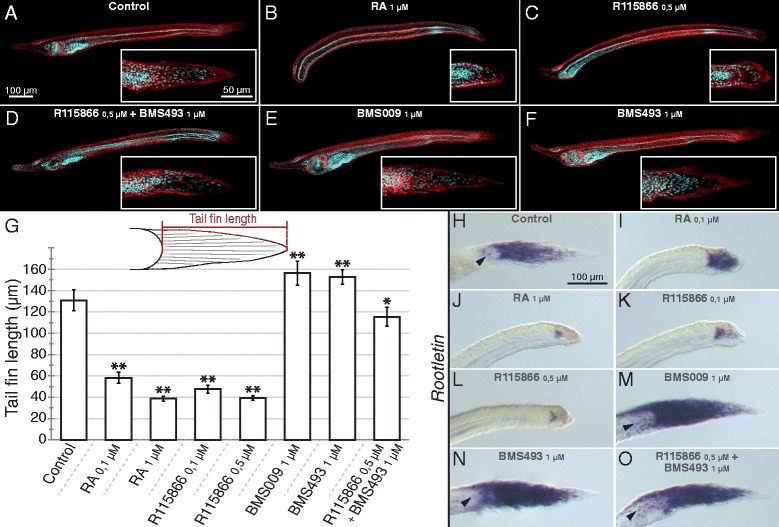
Effects of retinoic acid (RA) signaling alterations on the development of amphioxus. (a-f) Early amphioxus (Branchiostoma lanceolatum) larvae at 60 h of development with anterior to the left and the dorsal side up are shown as maximal projections of confocal microscopy scans displaying auto-fluorescence (in red) and Hoechst nuclear staining (in cyan). Magnifications of the tail region are boxed, with anterior to the left and the dorsal side up. Scale bars are 100 μm for the larvae and 50 μm for the magnified tail fins. (g) Tail fin length measurements following pharmacological treatments. The length of the tail fin of B. lanceolatum larvae at 60 h of development was measured, as indicated in the schematics. The graph shows the tail fin lengths ± standard deviation, with n = 15, for the different treatment conditions indicated. Tail fin lengths that are significantly different from the DMSO control are indicated as “*”, for a cutoff value of p = 0,005, and as “**”, for a cutoff value of p = 0,001. (h-o) Rootletin expression in the tail fin of early B. lanceolatum larvae at 60 h of development, with anterior to the left and the dorsal side up. Arrowheads in (h, m-o) indicate expression in lateral ectodermal cells. Pharmacological treatments are as indicated. Scale bars are 100 μm
Importantly, these pharmacology-based experiments revealed a previously undescribed role of RA signaling in developmental patterning of the amphioxus larval tail fin, a finding that is consistent with localized expression of CYP26-2 in the amphioxus tail fin ectoderm. At 60 h of development, amphioxus larvae are characterized by an ectodermal tail fin that is pointy in shape and on average 130,2 μm long (Fig. 3a and g). When amphioxus embryos were treated with RA at the gastrula stage, the resulting tail fins of 60-h larvae are round and significantly shorter. These effects were observable with exogenous treatments of both 0,1 μM and 1 μM RA (Fig. 3b and g). Similar results were obtained with R115866 treatments at 0,1 μM and 0,5 μM (Fig. 3c and g). Conversely, RAR antagonist treatments, with either BMS009 or BMS493, had the inverse effect: the tail fin becomes pointier in shape and is slightly elongated (Fig. 3e-g). Co-treatment of 0,5 μM R115866 with 1 μM BMS493 led to a partial rescue of the tail fin phenotype with an almost normal shape and a slight reduction of the overall length (Fig. 3d and g).
It has previously been shown that the amphioxus larval tail fin is composed of columnar epidermal cells that contain a large ciliary rootlet [49, 50] and that RA signaling promotes tail regression in late, pre-metamorphic B. floridae larvae by downregulating the gene encoding the main component of the ciliary rootlet: the protein Rootletin [51]. However, the regulation of Rootletin expression by RA signaling in the tail fin of early B. floridae larvae has not yet been reported. Given the effects on early tail fin formation we observed in B. lanceolatum in response to the alteration of endogenous RA signaling levels, we decided to investigate the patterns of Rootletin expression in 60-h B. lanceolatum larvae following the pharmacological treatment regimes detailed above. As previously described for late B. floridae larvae [51], Rootletin is expressed in the basal compartment of the columnar tail fin cells in 60-h B. lanceolatum larvae (Fig. 3h). At this developmental stage, the gene is further detectable in a small number of lateral ectodermal cells (Fig. 3h).
While treatment with 0,1 μM RA resulted in a marked reduction of Rootletin expression concomitant with an apical compaction of the columnar tail fin cells (Fig. 3i), 1 μM RA very strongly restricted the Rootletin expression domain (Fig. 3j). The effect on tail fin development of either 0,1 μM or 0,5 μM of the CYP26 inhibitor R115866 was similar to that of exogenous RA and the R115866-treated larvae were thus generally characterized by a significant reduction of Rootletin expression (Fig. 3k and l). The RAR antagonists BMS009 and BMS493 led to an apical expansion of Rootletin expression as well as to an increase of the overall length of the tail fin (Fig. 3m and n). Furthermore, the RAR antagonist treatments induced Rootletin expression in additional lateral ectodermal cells not directly associated with the tail fin (Fig. 3m and n). Intriguingly, while co-treatments of 0,5 μM R115866 and 1 μM BMS493 restored a shortened tail fin with almost normal shape, expression of Rootletin remained expanded into the apical territory of the tail fin and detectable in lateral ectodermal cells (Fig. 3o). Altogether, these observations suggest that the formation of the ectodermal tail fin in B. lanceolatum is dependent of RA signaling, with CYP26-dependent degradation playing an important role in fine tuning of endogenous RA signaling levels to ensure proper tail fin outgrowth.
CYP26-1 and CYP26-3 are highly responsive to RA and specifically upregulated to avoid teratogenic effects of RA
In vertebrates, CYP26 enzymes are known to function locally to reduce RA levels hence protecting target tissues from RA teratogenesis. In this context, the regulation of CYP26 activity is mediated, at least in part, by RA signaling, which very dynamically up- or down-regulates CYP26 expression in a tissue-dependent context [18]. To obtain insights into the regulation of CYP26 genes by RA signaling in B. lanceolatum, we performed quantitative real time PCR (qPCR) analyses on amphioxus embryos at two developmental stages (mid neurula and early larva) that have been treated, at the early gastrula stage, with either 1 μM RA or 1 μM of the RAR antagonist BMS009. The qPCR experiments assessed the changes in relative expression of the three B. lanceolatum CYP26 genes normalized by RAR expression in control embryos (Fig. 4a and b).
Fig. 4.
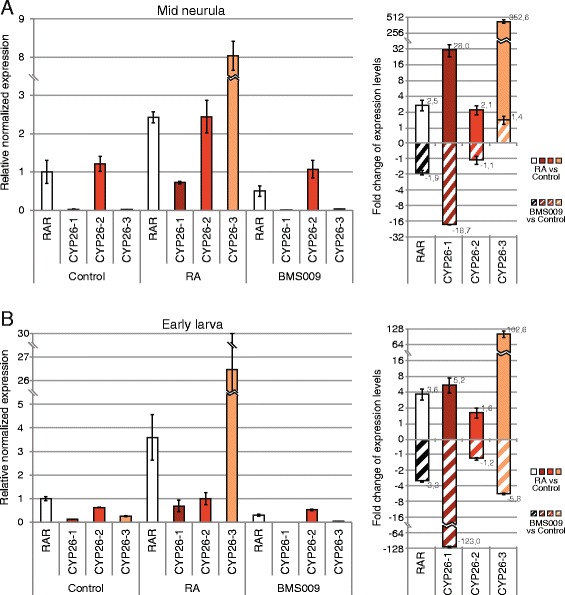
Quantitative changes of amphioxus CYP26 expression in response to retinoic acid (RA) signaling alterations. Expression of CYP26-1, CYP26-2, and CYP26-3 in amphioxus (Branchiostoma lanceolatum) was assessed by quantitative real time PCR (qPCR) at two developmental stages (mid neurula at 20 h of development and early larva at 48 h of development). RAR expression was also established and used as a normalized positive control. (a) Relative normalized expression (left panel) and fold change of expression levels (right panel) at the mid neurula stage, using a Log2 scale. (b) Relative normalized expression (left panel) and fold change of expression levels (right panel) at the early larval stage, using a Log2 scale
Through these analyses, we found that the expression levels of CYP26-1 and CYP26-3 in control embryos were very low when compared to CYP26-2 (about 45,0 to 50,0 times less in mid neurulae and 2,5 to 5,0 times less in early larvae) (Fig. 4a and b). RA treatments very significantly increased the expression of both CYP26-1 and CYP26-3 at the mid neurula stage (an average of 28 and 352,6 fold, respectively), while CYP26-2 levels increased merely by about 2,1 fold (Fig. 4a). In contrast, when embryos were treated with the RAR antagonist BMS009, CYP26-1 levels decreased by an average of 18,7 fold in mid neurulae, while the overall transcription of CYP26-2 and CYP26-3 remained relatively unchanged when compared to controls (a decrease of about 1,1 fold for CYP26-2 and an increase of about 1,4 fold for CYP26-3) (Fig. 4a). In early larvae, RA treatment also significantly increased the expression levels of CYP26-1 and CYP26-3 (by an average of 5,2 and 102,6 fold, respectively), while that of CYP26-2 increased only by about 1,6 fold (Fig. 4b). The RAR antagonist BMS009 had the opposite effect and strongly decreased CYP26-1 levels by about 123,0 fold and CYP26-3 levels by an average of 5,8 fold (Fig. 4b). In contrast, CYP26-2 expression dropped by only about 1,2 fold (Fig. 4b). These results indicate that, although CYP26-2 is the amphioxus CYP26 gene most strongly and broadly expressed during embryogenesis, CYP26-1 and CYP26-3 are much more reactive to alterations of endogenous RA signaling levels.
Following this quantification, we next assessed, by ISH, the developmental expression profiles of amphioxus CYP26 genes upon RA signaling-altering pharmacological treatments. Using the same developmental stages as for the qPCR analyses (mid neurulae and early larvae), we found that the upregulation of CYP26-1 expression upon RA treatment was not uniform (Fig. 5a-f). At the mid neurula stage, CYP26-1 was most strongly induced in the anterior half of the embryo in all tissue layers (i.e. in CNS, ectoderm, mesoderm, and endoderm), all along the CNS as well as in the posterior tip of the embryo in all tissue layers excepting the mesoderm (Fig. 5a and b). In early larvae, the gene was upregulated in all tissue layers of the anterior half of the animal, but largely undetectable posteriorly, except in a few cells in the ectoderm (Fig. 5d and e). As expected from the qPCR experiments, expression of CYP26-1 in embryos and larvae treated with the RAR antagonist BMS009 was very inconspicuous, but, after an extended coloration step, was nonetheless detectable in mesodermal tissues (Fig. 5c and f).
Fig. 5.
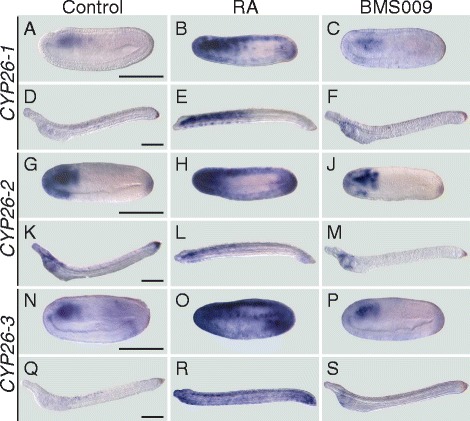
Spatial changes of amphioxus CYP26 expression in response to retinoic acid (RA) signaling alterations. Whole mount in situ hybridization experiments were carried out for CYP26-1 (a-f), CYP26-2 (g-m), and CYP26-3 (n-s) on amphioxus (Branchiostoma lanceolatum) embryos treated at the late blastula stage with either DMSO (as control), all-trans RA (1 μM) or RAR antagonist BMS009 (1 μM). The BMS009-treated embryo in (c) and larva in (f) were subjected to an extended coloration period. Mid neurula embryos at 20 h of development (a-c, g-j, n-p) and early larvae at 48 h of development (d-f, k-m, q-s) are shown as lateral views with anterior to the left and the dorsal side up. Scale bars are 100 μm
Treatment effects were similar, but not identical, for CYP26-2. At the mid neurula stage, RA strongly induced CYP26-2 anteriorly and posteriorly in all tissue layers as well as all along the CNS and ectoderm (Fig. 5h). However, following RA treatment, CYP26-2 expression was more conspicuous anteriorly and posteriorly in the embryo, when compared to that of CYP26-1. In contrast, in early larvae the effects of RA treatments were less pronounced for CYP26-2 than for CYP26-1. Chiefly, CYP26-2 expression expanded slightly in the mesoderm and ectoderm in the anterior half of the animal (Fig. 5k and l). Of note, the reduction of CYP26-2 expression in the anterior endoderm of RA-treated larvae was due to the absence of pharyngeal structures normally expressing the gene [43]. Treatments with the RAR antagonist BMS009 generally weakened the CYP26-2 signal, most noticeably in the CNS, the mesoderm, and both the anterior and posterior ectoderm (Fig. 5j and m).
Finally, the expression of CYP26-3 was also very strongly expanded by RA at mid neurula and early larva stages, even more strongly than that of CYP26-1 with a general expansion of the staining observed throughout the embryo at both stages (Fig. 5n and o). Following RA treatment, expression of the gene was induced anteriorly and posteriorly in all tissue layers, most conspicuously in the center of the embryo in the CNS, ectoderm, and mesoderm (Fig. 5n and o). By the early larval stage, CYP26-3 remained generally upregulated throughout the animal (Fig. 5q and r), which contrasts with a more restricted distribution of CYP26-1 and CYP26-2 transcripts at this stage of development in response to RA treatments (Fig. 5e and l). In embryos and larvae treated with the RAR antagonist BMS009, the CYP26-3 signal was very weak and chiefly limited to mesodermal tissues (Fig. 5p and s). Altogether, these results show that treatments with RA and RAR antagonist induce similar, but not identical, tissue-specific responses of amphioxus CYP26-1, CYP26-2, and CYP26-3, thereby supporting the notion that each one of the three CYP26 genes is required for a distinctive set of developmental functions.
RA regulates CYP26-1, CYP26-2, and CYP26-3 directly and via evolutionary conserved functional RAREs present in the CYP26 cluster
To assess, whether the observed effects of RA and RAR antagonist on amphioxus CYP26 expression were mediated directly by RAR/RXR heterodimers, we first carried out pharmacological treatments in the presence of puromycin, a compound that efficiently blocks de novo protein synthesis in developing amphioxus [44]. Amphioxus mid neurulae were hence treated with puromycin for 5 min prior to adding RA or the RAR antagonist BMS009. The embryos were then sampled 1 h later and, following RNA extraction, the expression levels of CYP26-1, CYP26-2, and CYP26-3 were determined by qPCR (Fig. 6). The results showed that, in the presence of puromycin, RA treatments significantly upregulated expression of both CYP26-1 and CYP26-3 (by about 5,8 fold and 5,9 fold, respectively), while that of CYP26-2 increased more modestly, by about 1,6 fold (Fig. 6). The RAR antagonist BMS009 had the inverse effect, reducing CYP26-1 and CYP26-3 levels by about 1,8 fold and 2,9 fold, respectively. In contrast, CYP26-2 expression stayed relatively stable, decreasing only by about 1,1 fold (Fig. 6). Altogether, these results are consistent with the effects of RA pharmacology on the expression of CYP26 genes described above. Furthermore, they indicate that the responses to RA and RAR antagonist treatments of all three amphioxus CYP26 genes do not require de novo protein synthesis, thereby implying that they must be mediated directly by RAR/RXR protein heterodimers present in the target tissues.
Fig. 6.
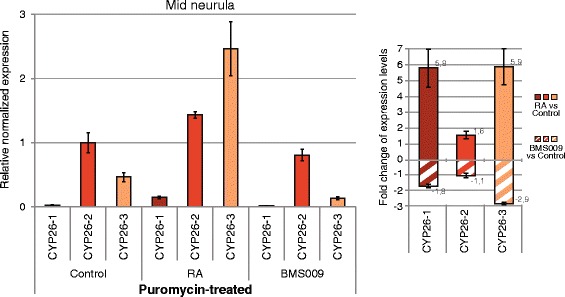
Dynamics of amphioxus CYP26 gene expression following protein synthesis inhibition and retinoid treatments. Expression of CYP26-1, CYP26-2, and CYP26-3 was assessed in amphioxus (Branchiostoma lanceolatum) by quantitative real time PCR (qPCR) at the mid neurula stage (at 20 h of development) following protein synthesis inhibition by puromycin treatment (200 μg/ml) at 19 h of development and subsequent treatment with DMSO (as control), all-trans RA (1 μM) or RAR antagonist BMS009 (1 μM) for 1 h. Relative normalized expression (left panel) and fold change of expression levels (right panel) relative to the controls are shown
In order to obtain deeper insights into the mechanisms of the direct regulation of CYP26-1, CYP26-2, and CYP26-3 by RA signaling we then investigated the genomic environment of CYP26 genes in the three amphioxus species with available genomes: B. floridae [36, 37], B. belcheri [52], and B. lanceolatum [53]. Our analyses allowed us to reconstruct the entire genomic clusters for the three amphioxus CYP26 genes, previously described only for B. floridae [29] (Fig. 7). Although variable in size (about 85 Kbp in B. floridae, 56 Kbp in B. belcheri, and 79 Kbp in B. lanceolatum from the start codon of CYP26-1 to the stop codon of CYP26-3), the order of the three CYP26 genes within the continuous clusters is conserved between the three amphioxus species, lending further support to the notion that these genes originated by tandem duplications from a single ancestral CYP26 gene at the base of the cephalochordates. Making use of the complete sequences of the three amphioxus CYP26 clusters, we further searched systematically for conserved RAREs in the vicinity of the CYP26-1, CYP26-2, and CYP26-3 genes. This in silico survey included a genomic region encompassing 20 Kbp upstream of the CYP26-1 start codon and 20 Kbp downstream of CYP26-3 stop codon (Fig. 7). In total, we identified 38 candidate RAREs in B. floridae, 30 in B. belcheri, and 21 in B. lanceolatum. Of these, 16 RAREs were conserved between all three amphioxus species, both in terms of DR motif sequences and their relative position within the CYP26 cluster (Fig. 7b and Additional file 4).
Fig. 7.
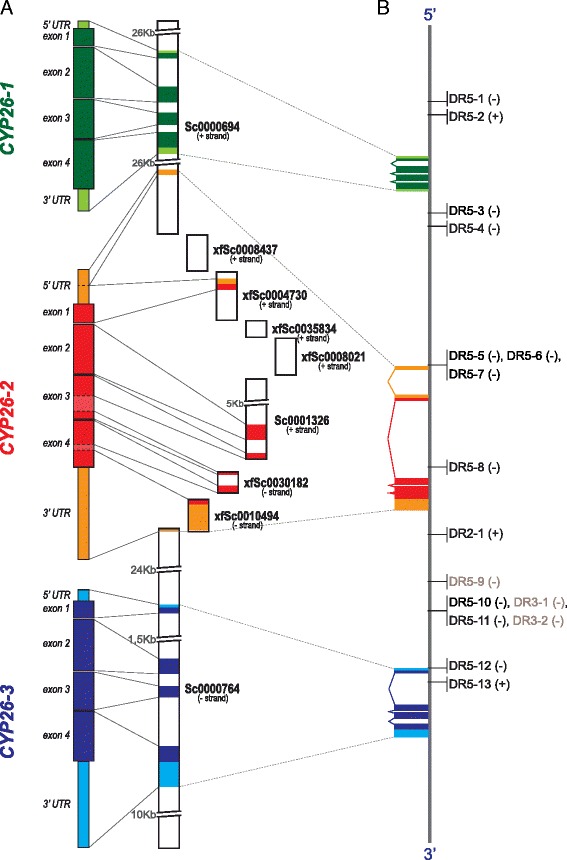
Organization and distribution of conserved retinoic acid response elements (RAREs) in the amphioxus CYP26 cluster. (a) Schematic representation of the experimentally verified amphioxus (Branchiostoma lanceolatum) CYP26-1, CYP26-2, and CYP26-3 gene sequences, including the 5′ and 3′ untranslated regions (UTRs) and the exons of the coding region, as well as of the corresponding scaffolds of the B. lanceolatum genome that cover the complete CYP26 cluster and thus also include intronic and intergenic regions. The names of the B. lanceolatum genome scaffolds are indicated, as is their orientation relative to the CYP26 cluster (+/− strand). (b) Distribution of conserved amphioxus RAREs in the B. lanceolatum CYP26 cluster. The orientation of each RARE is indicated relative to the CYP26 cluster (+/−). RAREs recognized and bound by the B. lanceolatum RAR/RXR heterodimer in vitro are indicated in black, RAREs that do not associate with the B. lanceolatum RAR/RXR heterodimer in vitro are shown in grey. The RARE sequences are given in Additional file 4 and the in vitro assays are shown in Fig. 8
These conserved amphioxus CYP26 RAREs, i.e. 13 DR5, 1 DR2, and 2 DR3 elements, were subsequently used in EMSA analyses to investigate their capacity, in vitro, to interact with the B. lanceolatum RAR/RXR heterodimer. Note that, even though DR3 elements are not considered as common RAREs, they were included in this survey. The results showed that the DR2 element and 12 of the 13 DR5 elements can be recognized and bound by the B. lanceolatum RAR/RXR heterodimer (Figs. 7b and 8), indicating that most of these RAREs might be functional in vivo. Furthermore, the consensus signature of the in vitro validated amphioxus DR5 elements [(A/G)G(G/T)T(C/G)A NNN(A/G)(A/C/G) (A/G)G(G/T)(T/A)CA] is very similar to the classical vertebrate DR5 signature [(A/G)G(G/T)TCA (N)5 (A/G)G(G/T)TCA] (Additional file 4), suggesting that, as in vertebrates [54, 55], amphioxus CYP26 genes are regulated directly by RA signaling via RAREs located in close vicinity of the open reading frames (ORFs).
Fig. 8.
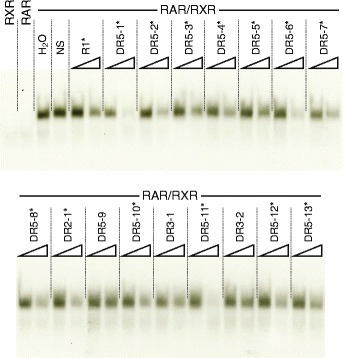
In vitro validation of putative retinoic acid response elements (RAREs) in the amphioxus CYP26 cluster. Using in vitro synthesized Branchiostoma lanceolatum RAR and RXR proteins, the binding of the cognate RAR/RXR heterodimer to putative RAREs identified in the amphioxus CYP26 cluster was assessed. The mouse CYP26A1 DR5 sequence (R1) [26] (Table 1) was used as 32P-radiolabeled double-stranded probe. The RXR and RAR lanes respectively contained only in vitro synthesized RXR or RAR proteins, together with the radiolabeled probe. The H20 (water) lane contained radiolabeled probe as well as RAR and RXR proteins. The NS (non-specific) lane contained a non-specific and unlabeled double-stranded DR element in 100-fold molar excess relative to the radiolabeled probe as well as RAR and RXR protein. All other lanes contained one of the 16 putative amphioxus DR elements (for nomenclature explanations see Fig. 7b) at 10-fold (left lane) or 100-fold (right lane) molar excess, along with RAR and RXR protein and the radiolabeled mouse R1 element, to test the binding specificity of the putative amphioxus RAREs. A black asterisk indicates that a given DR element can be recognized by the B. lanceolatum RAR/RXR heterodimer, as it outcompetes binding to the mouse R1 element
Furthermore, comparisons of the conserved and validated amphioxus CYP26 RAREs with functional RAREs associated with vertebrate CYP26 genes revealed that the amphioxus DR5-6 sequence located upstream of CYP26-2 (Fig. 7b and Additional file 4) is identical in sequence to a DR5 RARE located upstream of vertebrate CYP26A1 genes [AGTTCA (N)5 AGTTCA] [26, 54]. To verify, whether this DR5 RARE motif is a chordate innovation or an ancestral signature of bilaterian CYP26 genes, we screened the genomic regions surrounding CYP26 genes in the annelid C. teleta, the mollusk L. gigantea, the echinoderm S. purpuratus, the hemichordate S. kowalevskii, and three vertebrates (Takifugu rubripes, Mus musculus, and Homo sapiens). While no DR5 RARE with a similar motif could be identified in the annelid C. teleta and the mollusk L. gigantea, the conserved DR5 RARE was recovered in all three vertebrate species as well as in the hemichordate S. kowalevskii. Intriguingly, in the echinoderm S. purpuratus, instead of a DR5 RARE, we found a DR2 RARE with two similar DR sequences [AGTTCA] in an inverse orientation relative to the conserved DR5 RAREs in other species (Table 1). Of note, the conserved amphioxus DR5 RARE is located significantly further away from the CYP26 start codon than in the other studied species. Although the biological significance of this finding still remains to be explored, these results nonetheless suggest that the direct control of CYP26 expression by RA signaling, mediated at least in part by a conserved DR5 RARE, is an ancestral feature that was most likely absent in the last common ancestor of all bilaterians and that was thus only subsequently acquired at the base of the deuterostomes.
Table 1.
Conserved retinoic acid response elements (RAREs) with the characteristic sequence signature AGTTCA(N)5AGTTCA identified in the vicinity of bilaterian CYP26 genes
| Species | RARE name | Orientation relative to the CYP26 gene | Sequence (5′–3′) | Upstream of | Distance to start codon | Reference |
|---|---|---|---|---|---|---|
| S.p. | + | AGTTCAATAGTTCA | CYP26 | 2503 bp | This study | |
| S.k. | - | AGTTCATACCCAGTTCA | CYP26a | 137 bp | This study | |
| B.f. | DR5-6 | - | AGTTCAACAAAAGTTCA | CYP26-2 | 6405 bp | This study |
| B.b. | DR5-6 | - | AGTTCAACAAAAGTTCA | CYP26-2 | 6740 bp | This study |
| B.l. | DR5-6 | - | AGTTCAACAAAAGTTCA | CYP26-2 | 4083 bp | This study |
| H.s. | R1 | - | AGTTCACCCAAAGTTCA | CYP26A1 | 132 bp | [26] |
| M.m. | R1 | - | AGTTCACCCAAAGTTCA | CYP26A1 | 134 bp | [26] |
| D.r. | R1 | - | AGTTCACACAAAGTTCA | CYP26A1 | 166 bp | [26, 54] |
| R2 | - | AGTTCAAGGATAGTTCA | CYP26A1 | 1963 bp | ||
| T.r. | R1 | - | AGTTCATTCAAAGTTCA | CYP26A1 | 103 bp | This study |
Species name abbreviations: B.b. Branchiostoma belcheri, B.f. Branchiostoma floridae, B.l. Branchiostoma lanceolatum, D.r. Danio rerio, H.s. Homo sapiens, M.m. Mus musculus, S.k. Saccoglossus kowalevskii, S.p. Strongylocentrotus purpuratus, T.r. Takifugu rubripes
Discussion
CYP26 genes and their evolutionary involvement in RA-dependent A-P patterning in chordates
In this study, we assessed the developmental expression patterns and the biological functions of the three CYP26 duplicates from the European amphioxus B. lanceolatum. Globally, our results suggest a sub-functionalization of these genes. Of the three genes, CYP26-1 and CYP26-3 are characterized by weak and disperse expression patterns, while CYP26-2 displays a dynamic, tissue-specific pattern along the A-P axis. Considering that A-P patterning in early stages of amphioxus development is dependent on RA signaling [42, 44], the expression profile of CYP26-2, at very distinct positions along the A-P axis of the gastrula and early neurula, is suggestive of a functional role for this gene in this RA-dependent patterning process. This notion is further supported by the results obtained by pharmacological inhibition of CYP26 action during gastrulation, which yielded embryos and larvae that resemble those treated with exogenous RA, displaying severe A-P patterning defects [43]. Thus, we propose that CYP26 enzymes assume two main functions during amphioxus development (Fig. 9), which are equivalent to those observed in vertebrates: (1) the mediation of RA-dependent developmental patterning and (2) the protection against fluctuations of RA levels. Due to its conspicuous and tissue-specific expression during development, we hypothesize that the former is mainly assumed by CYP26-2, while the latter is dependent on the activity of CYP26-1 and CYP26-3.
Fig. 9.
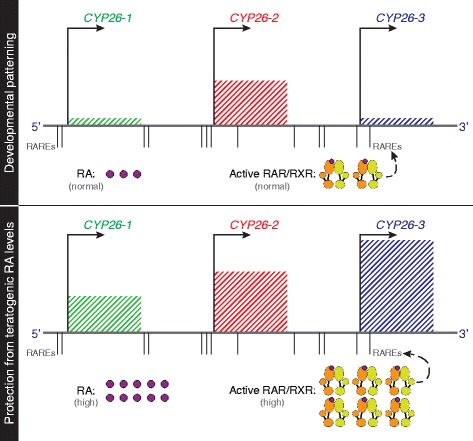
Model for regulation and function of CYP26 genes in amphioxus. The position of the functionally validated retinoic acid response elements (RAREs) within the amphioxus CYP26 cluster is indicated relative to the start codons of the three amphioxus CYP26 genes (CYP26-1, CYP26-2, and CYP26-3). The relative expression of each of the genes is indicated during normal development (normal levels of RA and active RAR/RXR receptor heterodimers) as well as during exposure to RA (high levels of RA and active RAR/RXR receptor heterodimers), hence highlighting the dominance of CYP26-2 during normal development and the importance of CYP26-1 and CYP26-3 for protection against RA teratogenesis
During early development, the expression domains of CYP26-2 along the A-P axis are inversely correlated with those of Hox1 and HNF3-1, both of which are direct targets of RA signaling in amphioxus [44, 56]. The posterior expression limit of CYP26-2 in anterior tissues thus abuts the anterior border of both the Hox1 and HNF3-1 domains [44]. Concomitantly, the blastopore-associated expression of CYP26-2 seems to delineate the posterior limits of both Hox1 and HNF3-1 [44, 56]. Given that RAR expression is ubiquitous during gastrulation [38], the presence of CYP26-2 in defined domains along the A-P axis might thus be crucial for the creation of RA sinks to subdivide the developing embryo into zones with and without active RA signaling. This is reminiscent of the conserved expression and function of CYP26A1 during vertebrate gastrulation: CYP26A1 is expressed in the anterior neural ectoderm and functions to establish A-P boundaries in the developing CNS [18]. It does so by establishing an anterior sink for RA produced posteriorly in the paraxial mesoderm, hence creating a RA gradient along the A-P axis of the neuroectoderm [57, 58].
By the mid neurula stage, the CYP26-2 signal is present in the entire anterior CNS, with a posterior limit at the boundary between the first and second somite. This CYP26 expression is hence limited to a region homologous to the vertebrate forebrain and midbrain [59], which is considerably different from what is observed in the vertebrate CNS, where CYP26 genes are expressed in the developing hindbrain and are fundamental for its patterning along the A-P axis [18, 19]. In the amphioxus hindbrain homolog, RA signaling mediated by RAR/RXR has been shown to confer regional identity along the A-P axis by controlling the collinear expression of Hox genes [42]. However, our work suggests that, in contrast to the situation in vertebrates, this process does not involve the deployment of CYP26 genes in the amphioxus hindbrain homolog. Thus, while the early role for CYP26 in neural patterning was probably already present in the last common ancestor of amphioxus and vertebrates, a CYP26-dependent mechanism for subsequent hindbrain regionalization probably evolved in vertebrates, following the vertebrate-specific CYP26 duplications.
Following neurulation, the three amphioxus CYP26 genes are expressed in anterior mesoderm, most noticeably in the anterior-most somites. This is intriguing, as to date there is no convincing evidence for a requirement of RA signaling in amphioxus mesoderm development [41, 42, 60] and the inhibition of CYP26 function by pharmacological treatments does not yield a mesodermal phenotype [43]. These observations raise an important question: why are CYP26 genes expressed in the anterior amphioxus mesoderm, as they are in vertebrate head mesoderm [25], if they are not required for A-P patterning of this tissue? We speculate that the conspicuous expression of CYP26 in the anterior somites is an amphioxus innovation and is thus not comparable to the role of CYP26 in the vertebrate head mesoderm. Instead, the CYP26 genes might function in the anterior amphioxus mesoderm to establish a buffer zone between the CNS dorsally and the pharynx ventrally, which require distinct RA signaling cues for A-P regionalization [41, 42]. Although further analyses aiming, for example, at the visualization of in vivo RA signaling levels [58] will be required to test this hypothesis, it would nonetheless be interesting to assess, whether this mechanism for separating two gradient-based A-P patterning systems is being used more widely during development of small-sized embryos.
In the course of development, the blastopore-associated expression of amphioxus CYP26-2 becomes incorporated into the tail bud, a structure that, from the neurula stage on, plays a central role in posterior elongation of the embryo and larva and that is further characterized by the expression of tissue-specific marker genes, such as Wnt3 [61]. In vertebrates, both CYP26A1 and Wnt3a are also co-expressed in the developing tail bud, and CYP26A1 null mutants are characterized by a truncated tail and a spatial expression of Wnt3a that is abnormally restricted towards the midline of the posterior neural plate [62]. These observations suggest that, in the vertebrate tail bud, CYP26A1 functions to keep RA levels low to create a permissive environment required for the posterior elongation of the embryo [16, 63]. In contrast, in amphioxus, treatments with CYP26 inhibitor, exogenous RA or the RAR antagonist BMS009 do not affect expression of Wnt3 in the tail bud [41]. Importantly, these pharmacological treatments, albeit affecting tail fin development, also do not impact posterior elongation of the developing embryo [39, 60]. Together, these findings support the notion that CYP26 function, and more generally the RA signaling system, is not required for tail bud-driven body extension in amphioxus and that this role for RA likely evolved in the vertebrate lineage.
CYP26 activity is required for the development of the amphioxus tail fin
Previous studies in the Florida amphioxus, B. floridae, have shown that the administration of excess RA during gastrulation results in a small anus [39] and that the continuous administration of RA to B. floridae larvae leads to closure of the anus and regression of the tail fin [51]. Our data on the European amphioxus, B. lanceolatum, are consistent with these previous findings and further suggest that RA signaling is required for proper tail fin outgrowth. Given that CYP26-2 is the only B. lanceolatum CYP26 gene expressed in the posterior ectoderm, it is very likely responsible for fine-tuning endogenous RA signaling levels in this territory.
The amphioxus tail fin is established by columnar epidermal cells that contain a large ciliary rootlet [49, 50] and it has previously been shown that RA promotes the downregulation of a major component of this structure, the protein Rootletin [51]. This downregulation in turn is likely responsible for the induction of tail fin regression upon RA treatments in B. floridae larvae [51]. Our results of RA and CYP26 inhibitor treatments in B. lanceolatum are generally consistent with these previous observations, although we identified a major difference in the developmental timing of the involvement of RA signaling in tail fin outgrowth between the two amphioxus species. This fact is exemplified by the experimental setups required to obtain tail fin phenotypes. While in B. floridae RA-dependent tail fin malformations can only be obtained by continuous treatment of pre-metamorphic larvae with 6–8 gill slits [51], changes in tail fin morphology can be induced much earlier during B. lanceolatum development by treating in the course of gastrulation. This suggests that, albeit required for tail fin outgrowth in both amphioxus species, the attenuation of high RA signaling levels in the posterior ectoderm might take place earlier in B. lanceolatum than in B. floridae development. It is possible that this developmental difference is mediated by a delayed activation of the expression of CYP26-2 in B. floridae relative to B. lanceolatum, although additional work is required to support this claim. Altogether, these results indicate that amphioxus tail fin development requires low levels of RA signaling, which are maintained by CYP26 activity. When compared to the known functions of CYP26 in vertebrates, where cells expressing CYP26 enzymes are said to be effectively devoid of RA [63], amphioxus tail fin outgrowth might represent a rare example of a developmental process, where CYP26 is deployed to merely reduce, and not to completely eliminate, RA signaling levels.
Amphioxus CYP26 genes are highly responsive RA signaling targets
In vertebrates, it has been established that, in RA-sensitive tissues, RA induces CYP26 to generate a negative feedback loop that reduces the overall amount of available RA [18]. In mice, for example, expression of both CYP26A1 and CYP26B1 is upregulated upon RA treatment [9] and, at least for CYP26A1, this regulation is directly mediated by RAR/RXR binding to RAREs in the vicinity of the CYP26A1 gene [26]. In contrast, the vertebrate CYP26C1 gene is likely not contributing to this RA-dependent negative feedback system, as its expression is actually downregulated upon RA stimulation [9]. The data presented here suggest that expression of all three amphioxus CYP26 genes, which are lineage-specific duplicates and located in a single cluster in the genome, is positively regulated by RA signaling. Accordingly, the transcriptional regulation of B. lanceolatum CYP26-1, CYP26-2, and CYP26-3 is in all likelihood directly mediated by RAR/RXR heterodimers, given that one DR2-type sequence and 12 DR5-type elements within the CYP26 cluster are recognized and bound by the B. lanceolatum RAR/RXR heterodimer in vitro. Of these 13 in vitro validated RAREs, four DR5 elements are located around the CYP26-1 coding sequence, four DR5 and one DR2 are found in proximity of CYP26-2, and four DR5 elements are associated with the CYP26-3 gene. Although each of the three amphioxus CYP26 genes are likely to be directly regulated by RA signaling, the specific arrangement of RAREs relative to the CYP26 ORFs nonetheless suggests that the expression of each of the three genes is regulated independently by different sets of RAREs. Although this fundamental difference in the regulation of amphioxus and vertebrate CYP26 genes remains to be demonstrated mechanistically in vivo, it is nonetheless tempting to speculate that alterations in the regulation by RA have been key for redefining the functions of the different CYP26 genes following their duplication, hence leading to their genomic retention by sub-functionalization.
At least some of the RAREs within the amphioxus CYP26 cluster might serve as hubs for long-range regulation of gene expression from shared RAREs. Although in both amphioxus and vertebrates it has previously been shown that functional RAREs are generally located in the proximity of genes directly regulated by RA signaling [16, 64], long-range regulatory mechanisms of RA signaling have, for example, been implicated in the control of the rostral expansion of posterior Hoxb genes during mouse CNS development [65]. The in vivo validation of the RAREs located within the amphioxus CYP26 cluster will shed light on their contribution to short- and/or long-range transcriptional control mechanisms exerted by RA signaling. Our in silico analyses further revealed the presence, within the amphioxus CYP26 cluster, of two DR3 elements in close proximity of one of the in vitro validated DR5 elements. Although DR3 elements are generally not recognized by RAR/RXR heterodimers, they are bound by other nuclear receptors, such as vitamin D receptors [66]. The presence of DR3 elements within the amphioxus CYP26 cluster thus hints at the possibility that additional nuclear receptors are involved in the regulation of amphioxus CYP26 genes.
Interestingly, the consensus sequence of the in vitro validated amphioxus DR5 RAREs is identical to the classical vertebrate DR5 sequence consensus [(A/G)G(G/T)TCA (N)5 (A/G)G(G/T)TCA] [67], suggesting that the DNA binding properties of amphioxus and vertebrate RAR/RXR heterodimers are highly conserved. Along these lines, we found a similar, conserved DR5 RARE in the regulatory region of a hemichordate CYP26 gene as well as an equivalent DR2 RARE close to the CYP26 gene of a sea urchin. Whether these elements are recognized by RAR/RXR heterodimers in these two species is currently unknown. Their presence is nevertheless highly suggestive of a biological function [13], which thus indicates that direct regulation of CYP26 genes by RA signaling is an ancestral feature that was already present in the last common ancestor of all deuterostomes. In contrast, the lack of conserved RAREs in the regulatory regions of the CYP26 genes of lophotrochozoans, which generally encode RAR and RXR genes in their genomes [2], support the notion that the direct regulation of CYP26 transcription by RA signaling is not an ancestral feature of bilaterian animals. This hypothesis is further strengthened by the fact that RARs of gastropod mollusks are unable to bind RA and hence to activate transcription in its presence [68, 69].
The evolutionary history of CYP26 genes
In the animal kingdom, the basic molecular components of the RA machinery have previously been described in a wide variety of bilaterian animals, including both protostomes and deuterostomes [2, 45]. Genes encoding the RA degrading enzyme CYP26 have, for example, been identified in the genomes of priapulids, brachiopods, mollusks, annelids, sea urchins, hemichordates, cephalochordates, ascidian tunicates, and vertebrates, but not in those of nematodes and arthropods, suggesting a secondary loss of CYP26 subfamily genes in these two animal lineages [2, 46]. We also found evidence for possible secondary losses of CYP26 genes in different vertebrates, including the lamprey P. marinus (lacking CYP26A1), the chimera C. milii (lacking CYP26A1), the skate L. erinacea (lacking CYP26C1), the coelacanth Latimeria chalumnae (lacking CYP26C1), and the opossum Monodelphis domestica (lacking CYP26B1) (Additional files 1 and 2). Additional sequence information will be required to validate the absence of these genes from their respective genomes and to assess, at which point in evolution the confirmed gene losses occurred.
Our data further indicate that the evolutionary diversification of the vertebrate CYP26 genes was a highly complex process. Given the presence of at least one CYP26A1 and two CYP26B1/C1 genes in lampreys, the last common ancestor of cyclostomes and gnathostomes probably already possessed at least two CYP26 genes. Intriguingly, in the lamprey L. japonicum, CYP26A1 and one of the two CYP26B1/C1, CYP26B1/C1a, are physically linked in a tandem cluster in the genome (on scaffold 19) (Additional file 1), just like CYP26A1 and CYP26C1 in gnathostome genomes [70]. This tandem cluster thus very likely originated before the cyclostome-gnathostome split, probably by a tandem duplication event predating the vertebrate-specific WGD. The two linked CYP26 genes were then duplicated during the first WGD, which took place in the basal vertebrate lineage before the cyclostome-gnathostome split [71–73]. The subsequent loss of one of the duplicated CYP26A1 genes yielded the CYP26 complement of extant lampreys, such as L. japonicum: one CYP26A1 and two CYP26B1/C1 genes. In the gnathostome lineage, additional duplications [71–73] resulted in the diversification of CYP26B1 and CYP26C1 genes from the ancestral CYP26B1/C1. Although requiring additional scrutiny, this proposed scenario for vertebrate CYP26 diversification suggests that, if two rounds of WGD occurred in the course of vertebrate evolution, the first likely took place before and the second after the cyclostome-gnathostome split [71, 72]. Alternatively, a single, ancient WGD might have occurred before the cyclostome-gnathostome split and was followed by independent segmental duplications in the cyclostome and gnathostome lineages [73]. Interestingly, in teleost fish, which underwent an additional round of WGD [74], there are also only three CYP26 genes, one member of each gnathostome paralogy group (CYP26A1, CYP26B1, and CYP26C1), and synteny analyses have revealed that the teleost-specific CYP26 duplicates have been non-functionalized in the course of evolution, in accordance with the DDC model [30, 31, 70].
Our phylogenetic analyses also provided evidence that CYP26 genes underwent multiple duplication events, not only in vertebrates, but also in invertebrates, such as cephalochordates, ascidian tunicates, hemichordates, mollusks, annelids, brachiopods, and priapulids. Together, these results suggest that the ancestral bilaterian possessed a single CYP26 gene that was subjected to independent duplication events in different animal lineages. By correlating the timing of the lineage-specific duplications of invertebrate CYP26 genes with the geological timescale, we observed that the duplication events fall within three distinct time periods, i.e. the late Ordovician, the early Carboniferous, and the middle Permian/middle Triassic. Thus, the priapulid CYP26 duplication took place during the late Ordovician, the first cephalochordate duplication and that identified in the hemichordate S. kowalevskii in the early Carboniferous, and the second cephalochordate duplication and that in the hemichordate P.flava in the middle Permian/middle Triassic. During the late Ordovician, a period marked by a mass extinction of marine species [75], earth was characterized by a rising level of atmospheric O2 [76] and by vast shallow and warm continental seas [75]. These conditions were very favorable for the appearance of cyanobacterial mats at moderate depths of the water column [77, 78]. Similarly, during the early Carboniferous atmospheric O2 levels were rising [76, 79], and the fossil record accounts for one of the highest concentrations of calcified marine cyanobacteria [78]. The middle Permian/middle Triassic, in turn, saw the greatest biotic crisis in earth’s history [80], and the fossil record suggests that, as a result of environmental changes, cyanobacteria became one of the most abundant life forms in both shallow and deep water environments [81].
Extant cyanobacteria that are known to create massive blooms under the exact same environmental conditions include Trichodesmium, Anabaena, and Synechocystis [82, 83], all of which are also known to produce high levels of carotenoids and retinoids as anti-oxidative byproducts, which have been shown to induce teratogenic effects in the surrounding fauna [83–85]. The prevalence of cyanobacterial mats in marine environments during the late Ordovician, early Carboniferous, and the middle Permian/middle Triassic might thus explain why CYP26 duplications that occurred during these two geological periods were independently retained, by either neo- or sub-functionalization [30, 31], in the genomes of different marine animal lineages. The independent duplication of CYP26 genes might have increased the overall fitness of a given population by favoring individuals that were more efficient in buffering fluctuations of exogenous retinoid levels. Altogether, this finding represents an intriguing example of adaptive convergent evolution in response to environmental changes.
Conclusions
In the present study, we characterized the expression, function, and regulation of CYP26 genes during amphioxus development. Our data suggest that, despite the independent origins of the CYP26 gene repertoires in chordates, the CYP26 genes of cephalochordates and vertebrates convergently evolved similar developmental functions: RA-dependent patterning and homeostatic regulation of RA levels. Moreover, by comparing the regulatory regions of CYP26 genes in three amphioxus species with those from several different animal taxa, we identified a highly conserved, functional RARE, suggesting that negative feedback regulation of RA signaling is an evolutionary ancient mechanism for controlling endogenous RA levels. This mechanism of regulation was likely already present in the last common ancestor of all deuterostomes, but not in that of all bilaterians. Finally, the correlation between the timing of lineage-specific duplications of bilaterian CYP26 genes and major environmental changes in the geological record suggest that the evolutionary diversification of the CYP26 subfamily in bilaterians was strongly influenced by environmental pressures to buffer fluctuations of exogenous retinoid levels. In sum, this work thus sheds light on the evolution of the regulation of endogenous RA levels and establishes a framework for studying adaptive convergent evolutionary changes following gene duplication.
Methods
Amphioxus adult husbandry, embryo rearing, and pharmacological treatments
Sexually mature animals of the European amphioxus (Branchiostoma lanceolatum) were collected by dredging in Argelès-sur-Mer, France, and retrieved from the sand by sieving. The collected animals were split evenly into tanks, with about 10–15 animals per aquarium, males and females together. The water temperature in the aquaria was kept at 16–17 °C, and the animals were kept under a spring-like day/night period, with 14 h of light and 10 h of absolute darkness. Spawning was induced by a 36-h thermal shock at 23 °C, as previously described [86–88]. Following oocyte and sperm collection and in vitro fertilization, the embryos were raised in artificial seawater, in the dark, at 19 °C [89]. Pharmacological treatments of B. lanceolatum embryos were performed at the late blastula stage (6 h of development) with all-trans RA (at 0,1 μM and 1 μM) (Sigma-Aldrich, Saint-Quentin Fallavier, France), the RAR antagonists BMS009 (at 1 μM) or BMS493 (at 1 μM) (Sigma-Aldrich, Saint-Quentin Fallavier, France), the CYP26 inhibitor R115866 (at 0,5 μM and 0,1 μM) (provided by Janssen Research & Development, a division of Janssen Pharmaceutica NV, Beerse, Belgium) or with a combination of both BMS493 (at 1 μM) and R115866 (at 0,5 μM). All compounds were initially dissolved in dimethyl sulfoxide (DMSO) to create 1000X stock solutions and subsequently added to the embryo cultures in artificial seawater in a 1:1000 dilution to yield the respective final concentrations. As a control, embryos were treated in separate dishes with DMSO alone to a final dilution of 1:1000 [38, 39]. Puromycin (Sigma-Aldrich, Saint-Quentin Fallavier, France) treatments were performed at the mid neurula stage (19 h of development) by adding the compound to embryo cultures at a final concentration of 200 μg/ml. After 5 min of incubation, all-trans RA (1 μM), the RAR antagonist BMS009 (1 μM) or DMSO (at a dilution of 1:1000) were added, and, 1 h thereafter, the embryos were frozen for RNA extraction.
Sequence analyses and gene cloning
The B. lanceolatum CYP26-1, CYP26-2, and CYP26-3 sequences were obtained in silico from the B. lanceolatum genome by local BLAST using as template the sequences of B. floridae and B. belcheri CYP26 (Additional file 1). RNA was extracted from embryos at different developmental stages according to the established protocols [90] and cDNA was synthesized using the SuperScriptIII reverse transcription kit (Invitrogen, Cergy Pontoise, France). Complete coding sequences of B. lanceolatum CYP26-1, CYP26-2, and CYP26-3 were subsequently cloned by PCR using gene-specific primers containing the start and stop codons of the three CYP26 genes. RACE-PCR experiments using the SMARTer™ RACE cDNA amplification kit (Clontech, Saint-Germain-en-Laye, France) were then performed to amplify the 5′ and 3′ untranslated regions (UTRs) of B. lanceolatum CYP26-1, CYP26-2, and CYP26-3. The PCR and RACE-PCR products were cloned into the pCRII-TOPO vector (Invitrogen, Cergy Pontoise, France) and sequenced on both strands for validation. The B. lanceolatum CYP26-1, CYP26-2, and CYP26-3 sequences were deposited in GenBank and their accession numbers are as follows: CYP26-1 (KX118106), CYP26-2 (KX118108), and CYP26-3 (KX118107). Furthermore, a 2291-bp piece of the B. lanceolatum Rootletin gene was amplified by PCR, cloned into the pGEM-T Easy vector (Promega, Charbonnières-les-Bains, France), and verified by sequencing on both strands (GenBank accession number: KX118111), before being used as a marker for in situ hybridization (ISH) experiments.
Phylogenetic analyses
Phylogenetic trees of the CYP26 subfamily were calculated from amino acid and nucleotide sequences and the sequences included in the analysis are listed in Additional file 1, along with representative members of the CYP51 subfamily, which were used as outgroup. Nucleotide sequences were translated into amino acid sequences, aligned with Muscle as implemented in SeaView v4.5.4 [91], and refined by eye (Additional file 5). The final amino acid alignment was subsequently retransformed into the final nucleotide alignment (Additional file 6). The best-fit models of amino acid and nucleotide sequence evolution were selected based on the AIC score implemented, respectively, in ProtTtest 3 [92] and jModelTest v2.1 [93]. Molecular phylogenies were calculated with the Maximum Likelihood (ML) method using PhyML v3.1 [94] under the models selected by ProtTtest 3 (LG + I + G + F) and jModelTest v2.1 (GTR + I + G). In addition, Bayesian Inference (BI) analyses were performed using the program MrBayes v3.2.6 [95] and the same models. The robustness of each node was estimated by bootstrap analyses (in 1000 pseudoreplicates) using PhyML v3.1 for the ML tree and by posterior probability for the BI tree, with two Monte Carlo Markov Chain (MCMC) analyses run independently for 100 million generations and trees sampled every 1000 generations. The burn-in was determined with Tracer v1.6 [96], and the average standard deviation of split frequencies remained at <0.05 after the burn-in threshold. 10% and 50% of the trees were discarded, respectively, for the amino acid and nucleotide datasets. Consensus trees were visualized with Figtree v1.4 [97].
Comparative dating analysis
Based on the nucleotide dataset, the divergence dates of the CYP26 sequences were estimated with Beast v2.4 [98] using divergence times in million years ago (Mya) estimated with the calibration intervals proposed by Benton and colleagues and dos Reis and colleagues [99, 100]: origin of Vertebrata (457,5-636,1 Mya), Euarchontoglires (61,6-164,6 Mya), and Hemichordata (504,5-636,1 Mya) as well as the divergence between Holostei and Teleostei (250,0–331,1 Mya) and between Otocephala and Euteleostei (150,94–235 Mya). All calibration constraint sets were defined following a hard minimum, soft upper boundaries, and a lognormal prior [98]. Monte Carlo Markov Chain (MCMC) analyses were run on the nucleotide dataset for 100 million generations with trees sampled every 1000 generations [98]. Convergence of the calculations was verified and burn-in estimated with Tracer v1.6 [96]. The results of the MCMC run were sampled with LogCombiner and a burn-in of 30% [98]. The trees were combined into a maximum clade credibility tree using TreeAnnotator with an estimation of the mean node height and highest posterior density intervals fixed at 95% [98].
B. lanceolatum CYP26 cluster reconstitution and identification of putative RAREs
The full-length ORFs of B. lanceolatum CYP26-1, CYP26-2, and CYP26-3 were used as templates for local BLAST searches of the B. lanceolatum genome to identify scaffolds containing UTRs and coding regions of CYP26-1, CYP26-2, and CYP26-3. Sequences of genome regions not obtained by these BLAST approaches (hence corresponding to gaps in the B. lanceolatum CYP26 cluster) were subsequently identified by reciprocal BLAST searches of the B. lanceolatum genome sequence using short regions (less than 2 Kbp) of the B. floridae and B. belcheri CYP26 clusters. Putative RARE sequences were identified with an automated pipeline [51] using as input RAREs previously described as functional in amphioxus as well as all possible RARE combinations resulting from the canonical vertebrate RARE consensus: (A/G)G(G/T)TCA(N)0–9(A/G)G(G/T)TCA. Detailed sequence and scaffold information is provided in Additional files 4 and 7.
Multiple expectation maximization algorithm for motif elicitation (MEME) analysis
Multiple expectation maximization algorithm for motif elicitation (MEME) logos were calculated with MEME Suite v4.10.1 [101] using as input file the sequences of the in vitro validated B. lanceolatum DR5 RARE sequences including 5 nucleotides upstream and downstream of the element. The following settings were used to obtain the MEME logos: nmotifs = 2, minwidth = 15, maxwidth = 27.
Electrophoretic mobility shift assay (EMSA) experiments
The B. lanceolatum RAR and RXR coding sequences were amplified by PCR and cloned into the pGEM-T Easy vector (Promega, Charbonnières-les-Bains, France). Subcloning into the pCS2+ vector [102] was performed using introduced EcoRI and XhoI restriction sites. Following verification by sequencing on both strands of the RAR-pCS2+ and RXR-pCS2+ constructs (GenBank accession numbers: B. lanceolatum RAR, KX118109; B. lanceolatum RXR, KX118110), RAR and RXR proteins were produced by in vitro translation using the TNT coupled reticulocyte lysate system (Promega, Charbonnières-les-Bains, France). Electrophoretic mobility shift assays (EMSAs) were performed as previously described [69] using 4 μl of each receptor synthesized in vitro and 30–50 × 103 CPM of double-stranded oligonucleotide probe end-labeled with (γ-32P)ATP, incubated for 30 min on ice in a final volume of 20 μl of binding buffer: 20 mM Tris–HCl pH8, 50 mM KCl, 2 mM DTT, 25 mM MgCl2, 50 mM NaCl, 1 μg poly (dI-dC), and 10% glycerol. The samples were subsequently run on a 5% native acrylamide gel in 1X TAE for 2 h.
In situ hybridization (ISH), histology, imaging, and tail fin measurements
Antisense riboprobe synthesis, ISH, and Hoechst staining (Invitrogen, Cergy Pontoise, France) experiments were performed as previously described [103]. For ISH, Hoechst staining, and tail fin measurements, amphioxus (B. lanceolatum) embryos and larvae were fixed in 4% paraformaldehyde at different developmental stages, as previously described [103]. Following ISH, B. lanceolatum embryos and larvae were first photographed as whole mounts using Zeiss DIC (differential interference contrast) optics (Carl Zeiss SAS, Marly le Roi, France) and subsequently counterstained in Ponceau S (Sigma-Aldrich, Saint-Quentin Fallavier, France), embedded in Spurr’s resin (Sigma-Aldrich, Saint-Quentin Fallavier, France), and prepared as 3 μm sections for light microscopy observations and photography. Following Hoechst staining, B. lanceolatum larvae were embedded in Mowiol mounting medium (Sigma-Aldrich, Saint-Quentin Fallavier, France) overnight at 4 °C, before being imaged using a Leica TCS SP5 confocal microscope (Leica Microsystems SAS, Nanterre, France). ImageJ was subsequently used for image processing and for the creation of maximal projections [104]. For tail fin measurements, normal and treated B. lanceolatum larvae were photographed using Zeiss DIC optics (Carl Zeiss SAS, Marly le Roi, France). The length of the tail fin, defined as the distance between the anterior-most end of the tail fin ectoderm and the posterior-most tip of the tail fin (as represented in Fig. 3g), was subsequently measured using the measurement tool of ImageJ [104] and ultimately represented as mean ± standard deviation, with n = 15 for each treatment and control conditions. Student’s t-test was used to assess the statistical significance of the length differences measured in the treated specimens relative to the DMSO control.
Quantitative real time PCR (qPCR) assays
Quantitative real time PCR (qPCR) experiments were performed at two developmental stages (mid neurula at 20 h of development and early larva at 48 h of development). In addition to the puromycin-treated material described above, the cDNAs from DMSO treatment controls as well as from embryos treated at the late blastula stage (6 h of development) with either 1 μM all-trans RA or 1 μM of the RAR antagonist BMS009 were assayed on a MJ Research DNA Engine Opticon system (Bio-Rad, Marnes-la-Coquette, France) using the QuantiTect SYBR Green PCR reagent (Qiagen SAS, Courtaboeuf, France) and primers specific for B. lanceolatum CYP26-1, CYP26-2, CYP26-3, RAR, and 18S rRNA (Additional file 8). Based on the lack of response to the different pharmacological treatments assayed, 18S rRNA was selected as the reference for internal standardization of the starting quantity of RNA. Each qPCR experiment was performed in triplicates and the relative expression was normalized to RAR or CYP26-2 expression levels, in Figs. 4 and 6, respectively. Normalized expression levels are shown as ΔΔCT means ± standard deviation, with n = 3. Furthermore, fold change of expression relative to the control is represented as the mean of the ΔΔCT ratios of all possible combinations of the three replicates of a given condition over the three controls ± standard deviation, with n = 9.
Acknowledgements
The Observatoire Océanologique de Banyuls-sur-Mer in Banyuls-sur-Mer, France, graciously provided Branchiostoma lanceolatum adults and Hector Escriva kindly coordinated and supported the animal collections. This work was supported by the Branchiostoma lanceolatum genome consortium that provided access to the Branchiostoma lanceolatum genome sequence. The authors are indebted to Janssen Research & Development, a division of Janssen Pharmaceutica NV, for providing the CYP26 inhibitor. Furthermore, we would like to thank Laurent Gilletta and Sophie Collet for maintaining amphioxus adults, Elisabeth Zieger for help with confocal imaging and for commenting the manuscript, Nicolas Robert and Ferdinand Marlétaz for fruitful discussions as well as François Lahaye and Guy Lhomond for technical support.
Funding
João E. Carvalho is a FCT doctoral fellow (SFRH/BD/86878/2012). The experiments reported in this study were financed by a grant from the Agence Nationale de la Recherche (ANR-11-JSV2-002-01) and funds from the Réseau André Picard (ANR-11-IDEX-0004-02, Sorbonne Universities) to Michael Schubert.
Availability of data and materials
Supporting data for this article are included in the supplementary information files. Additional raw data are available from the authors upon request.
Authors’ contributions
JEC performed experiments, analyzed data, compiled figures, wrote, and revised the manuscript. MT and JC contributed to gene cloning and expression analyses. PC carried out the phylogenetic analyses and revised the manuscript. SA and ARDL produced and provided reagents. VL assisted in the writing of the manuscript. JCC assisted in the writing and editing of the manuscript. MS conceived the study, analyzed data, wrote, edited, and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- A-P
Anterior-posterior
- BI
Bayesian inference
- BLAST
Basic local alignment search tool
- BMS
Bristol-Myers Squibb
- cDNA
Complimentary deoxyribonucleic acid
- CNS
Central nervous system
- CYP26
Cytochrome P450 subfamily 26
- CYP51
Cytochrome P450 subfamily 51
- DDC
Duplication degeneration complementation
- DIC
Differential interference contrast
- DMSO
Dimethyl sulfoxide
- DR
Direct repeat
- EMSA
Electrophoretic mobility shift assay
- FGF
Fibroblast growth factor
- HNF
Hepatocyte nuclear factor
- Hox
Homeobox gene
- ISH
In situ hybridization
- Kbp
Kilo base pairs = 1000 base pairs
- MCMC
Monte Carlo Markov chain
- MEME
Multiple expectation maximization algorithm for motif elicitation
- ML
Maximum likelihood
- Mya
Million years ago
- ORF
Open reading frame
- qPCR
Quantitative real time polymerase chain reaction
- RA
Retinoic acid
- RACE
Rapid amplification of complimentary deoxyribonucleic acid ends
- RALDH
Retinaldehyde dehydrogenase
- RAR
Retinoic acid receptor
- RARE
Retinoic acid response element
- rRNA
Ribosomal ribonucleic acid
- RXR
Retinoid X receptor
- UTR
Untranslated region
- WGD
Whole genome duplication
- Wnt
Wingless-related integration site
- ΔΔCT
Delta delta threshold cycle
Additional files
Accession numbers of the CYP26 and CYP51 subfamily sequences used to calculate the phylogenetic trees and to carry out the dating analyses. Species names and database information are provided for each sequence. (XLSX 12 kb)
Phylogenetic tree of the CYP26 subfamily, with CYP51 as outgroup. Tree calculated with the Bayesian Inference (BI) and Maximum Likelihood (ML) methods based on 15 vertebrate species (42 CYP26 sequences), 1 tunicate species (2 CYP26 sequences), 3 cephalochordate species (9 CYP26 sequences), 3 ambulacrarian species (6 CYP26 sequences), 5 lophotrochozoan species (12 CYP26 sequences), and 1 ecdysozoan species (2 CYP26 sequences) (for details see Additional file 1). The robustness of each node was assessed by posterior probability (for the BI phylogeny) and bootstrap (for the ML phylogeny) analyses, which are indicated at each node: posterior probabilities (ranging from 0 to 1)/bootstrap percentages (ranging from 0 to 100). “--” indicates nodes where the ML tree did not recover the branching pattern of the BI tree or nodes with bootstrap support inferior to 50%. Branch lengths are representative of the amino acid substitution rate. Species name abbreviations: B.b., Branchiostoma belcheri (in blue); B.f., Branchiostoma floridae (in red); B.l., Branchiostoma lanceolatum (in green); C.g., Crassostrea gigas; C.i., Ciona intestinalis; C.m., Callorhinchus milii; C.t., Capitella teleta; D.r., Danio rerio; G.a., Gasterosteus aculeatus; G.g., Gallus gallus; H.s., Homo sapiens; L.a. Lingula anatina; L.c., Latimeria chalumnae; L.e., Leucoraja erinacea; L.g., Lottia gigantea; L.j., Lethenteron japonicum; L.o., Lepisosteus oculatus; M.d., Monodelphis domestica; M.m., Mus musculus; O.b., Octopus bimaculoides; O.l., Oryzias latipes; P.c., Priapulus caudatus; P.f., Ptychodera flava; P.m., Petromyzon marinus; S.k., Saccoglossus kowalevskii; S.p., Strongylocentrotus purpuratus; T.r., Takifugu rubripes; X.t., Xenopus tropicalis. (PDF 559 kb)
Comparative dating tree for the CYP26 subfamily. Divergence dates of the CYP26 sequences were estimated using fossil-based calibration intervals. Cladogram node dates represent estimation of the mean node height and the highest posterior density intervals fixed at 95% are shown in parentheses. Geological era abbreviations: Cam, Cambrian; Car, Carboniferous; Cre, Cretaceous; Dev, Devonian; Jur, Jurassic; Ord, Ordovician; Per, Permian; Sil, Silurian; Tri, Triassic. Species name abbreviations: B.b., Branchiostoma belcheri (in blue); B.f., Branchiostoma floridae (in red); B.l., Branchiostoma lanceolatum (in green); C.g., Crassostrea gigas; C.i., Ciona intestinalis; C.m., Callorhinchus milii; C.t., Capitella teleta; D.r., Danio rerio; G.a., Gasterosteus aculeatus; G.g., Gallus gallus; H.s., Homo sapiens; L.a. Lingula anatina; L.c., Latimeria chalumnae; L.e., Leucoraja erinacea; L.g., Lottia gigantea; L.j., Lethenteron japonicum; L.o., Lepisosteus oculatus; M.d., Monodelphis domestica; M.m., Mus musculus; O.b., Octopus bimaculoides; O.l., Oryzias latipes; P.c., Priapulus caudatus; P.f., Ptychodera flava; P.m., Petromyzon marinus; S.k., Saccoglossus kowalevskii; S.p., Strongylocentrotus purpuratus; T.r., Takifugu rubripes; X.t., Xenopus tropicalis. (PDF 271 kb)
Conserved retinoic acid response elements (RAREs) found in the Branchiostoma lanceolatum, Branchiostoma belcheri, and Branchiostoma floridae CYP26 clusters. B. lanceolatum RAREs recognized and bound by the B. lanceolatum RAR/RXR heterodimer in vitro (Fig. 8) are indicated in black, RAREs that do not associate with the B. lanceolatum RAR/RXR heterodimer in vitro are shown in grey. The in vitro validated B. lanceolatum RAREs composed of two direct repeats (DRs) and separated by a 5 nucleotide spacer (i.e. the validated B. lanceolatum DR5-type RAREs) were used to calculate a consensus B. lanceolatum DR5 RARE by multiple expectation maximization algorithm for motif elicitation (MEME) analysis. (XLSX 77 kb)
Amino acid alignment used for phylogenetic tree reconstruction analyses of the CYP26 subfamily. (NEX 46 kb)
DNA alignment used for comparative dating analyses of the CYP26 subfamily. (NEX 135 kb)
Species names, corresponding genome access, and scaffold information related to the sequences used as input for the in silico identification of conserved putative retinoic acid response elements (RAREs). (XLSX 9 kb)
Sequences of the oligonucleotides used as primers for quantitative real time PCR (qPCR) analyses of the indicated Branchiostoma lanceolatum genes. (XLSX 8 kb)
Contributor Information
João E. Carvalho, Email: carvalho@obs-vlfr.fr
Maria Theodosiou, Email: mtheodos2@gmail.com.
Jie Chen, Email: j_chen@shou.edu.cn.
Pascale Chevret, Email: pascale.chevret@univ-lyon1.fr.
Susana Alvarez, Email: sar@uvigo.es.
Angel R. De Lera, Email: qolera@uvigo.es
Vincent Laudet, Email: vincent.laudet@obs-banyuls.fr.
Jenifer C. Croce, Email: jeni.croce@obs-vlfr.fr
Michael Schubert, Email: Michael.Schubert@obs-vlfr.fr.
References
- 1.Blomhoff R, Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol. 2006;66:606–30. doi: 10.1002/neu.20242. [DOI] [PubMed] [Google Scholar]
- 2.Campo-Paysaa F, Marlétaz F, Laudet V, Schubert M. Retinoic acid signaling in development: tissue-specific functions and evolutionary origins. Genesis. 2008;46:640–56. doi: 10.1002/dvg.20444. [DOI] [PubMed] [Google Scholar]
- 3.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–31. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niederreither K, Dollé P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–53. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 5.Noy N. Between death and survival: retinoic acid in regulation of apoptosis. Annu Rev Nutr. 2010;30:201–17. doi: 10.1146/annurev.nutr.28.061807.155509. [DOI] [PubMed] [Google Scholar]
- 6.Theodosiou M, Laudet V, Schubert M. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell Mol Life Sci. 2010;67:1423–45. doi: 10.1007/s00018-010-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho JE, Schubert M. Retinoic acid: metabolism, developmental functions, and evolution. In: Dakshinamurti K, Dakshinamurti S, editors. Vitam-Bind Proteins Funct Consequences. Boca Raton: CRC Press; 2013. p. 1–30.
- 8.Dobbs-McAuliffe B, Zhao Q, Linney E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev. 2004;121:339–50. doi: 10.1016/j.mod.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Lee LMY, Leung C-Y, Tang WWC, Choi H-L, Leung Y-C, McCaffery PJ, et al. A paradoxical teratogenic mechanism for retinoic acid. Proc Natl Acad Sci U S A. 2012;109:13668–73. doi: 10.1073/pnas.1200872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilling TF, Nie Q, Lander AD. Dynamics and precision in retinoic acid morphogen gradients. Curr Opin Genet Dev. 2012;22:562–9. doi: 10.1016/j.gde.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Aniello E, Rydeen AB, Anderson JL, Mandal A, Waxman JS. Depletion of retinoic acid receptors initiates a novel positive feedback mechanism that promotes teratogenic increases in retinoic acid. PLoS Genet. 2013;9:e1003689. doi: 10.1371/journal.pgen.1003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rydeen A, Voisin N, D’Aniello E, Ravisankar P, Devignes C-S, Waxman JS. Excessive feedback of Cyp26a1 promotes cell non-autonomous loss of retinoic acid signaling. Dev Biol. 2015;405:47–55. doi: 10.1016/j.ydbio.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–808. doi: 10.1194/jlr.R100015-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–66. doi: 10.1016/0092-8674(91)90020-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross SA, McCaffery PJ, Drager UC, Luca LMD. Retinoids in embryonal development. Physiol Rev. 2000;80:1021–54. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 2015;16:110–23. doi: 10.1038/nrm3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topletz AR, Tripathy S, Foti RS, Shimshoni JA, Nelson WL, Isoherranen N. Induction of CYP26A1 by metabolites of retinoic acid: evidence that CYP26A1 is an important enzyme in the elimination of active retinoids. Mol Pharmacol. 2015;87:430–41. doi: 10.1124/mol.114.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White RJ, Schilling TF. How degrading: Cyp26s in hindbrain development. Dev Dyn. 2008;237:2775–2790. doi: 10.1002/dvdy.21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–87. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yashiro K, Zhao X, Uehara M, Yamashita K, Nishijima M, Nishino J, et al. Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev Cell. 2004;6:411–22. doi: 10.1016/S1534-5807(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 21.Uehara M, Yashiro K, Mamiya S, Nishino J, Chambon P, Dollé P, et al. CYP26A1 and CYP26C1 cooperatively regulate anterior–posterior patterning of the developing brain and the production of migratory cranial neural crest cells in the mouse. Dev Biol. 2007;302:399–411. doi: 10.1016/j.ydbio.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, Duester G. Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development. 2014;141:2972–7. doi: 10.1242/dev.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Cunningham TJ, Duester G. Nuclear receptor corepressors Ncor1 and Ncor2 (Smrt) are required for retinoic acid-dependent repression of Fgf8 during somitogenesis. Dev Biol. 2016;418:204–15. doi: 10.1016/j.ydbio.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahl MB, Deng C, Lewandoski M, Pourquié O. FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development. 2007;134:4033–41. doi: 10.1242/dev.009167. [DOI] [PubMed] [Google Scholar]
- 25.Bothe I, Tenin G, Oseni A, Dietrich S. Dynamic control of head mesoderm patterning. Development. 2011;138:2807–21. doi: 10.1242/dev.062737. [DOI] [PubMed] [Google Scholar]
- 26.Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–97. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- 27.Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M. Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem J. 2005;392:241. doi: 10.1042/BJ20050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escriva H, Bertrand S, Germain P, Robinson-Rechavi M, Umbhauer M, Cartry J, et al. Neofunctionalization in vertebrates: the example of retinoic acid receptors. PLOS Genet. 2006;2:e102. [DOI] [PMC free article] [PubMed]
- 29.Albalat R, Brunet F, Laudet V, Schubert M. Evolution of retinoid and steroid signaling: vertebrate diversification from an amphioxus perspective. Genome Biol Evol. 2011;3:985–1005. doi: 10.1093/gbe/evr084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurles M. Gene duplication: the genomic trade in spare parts. PLoS Biol. 2004;2:e206. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacCarthy T, Bergman A. The limits of subfunctionalization. BMC Evol Biol. 2007;7:213. doi: 10.1186/1471-2148-7-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet P-L, et al. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007;3:e188. doi: 10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimeld SM, Holland ND. Amphioxus molecular biology: insights into vertebrate evolution and developmental mechanisms. Can J Zool. 2005;83:90–100. doi: 10.1139/z04-155. [DOI] [Google Scholar]
- 35.Bertrand S, Escriva H. Evolutionary crossroads in developmental biology: amphioxus. Development. 2011;138:4819–30. doi: 10.1242/dev.066720. [DOI] [PubMed] [Google Scholar]
- 36.Holland LZ, Albalat R, Azumi K, Benito-Gutiérrez È, Blow MJ, Bronner-Fraser M, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–11. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–71. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 38.Escriva H, Holland ND, Gronemeyer H, Laudet V, Holland LZ. The retinoic acid signaling pathway regulates anterior/posterior patterning in the nerve cord and pharynx of amphioxus, a chordate lacking neural crest. Development. 2002;129:2905–16. doi: 10.1242/dev.129.12.2905. [DOI] [PubMed] [Google Scholar]
- 39.Holland LZ, Holland ND. Expression of AmphiHox-1 and AmphiPax-1 in amphioxus embryos treated with retinoic acid: insights into evolution and patterning of the chordate nerve cord and pharynx. Development. 1996;122:1829–38. doi: 10.1242/dev.122.6.1829. [DOI] [PubMed] [Google Scholar]
- 40.Schubert M, Holland ND, Escriva H, Holland LZ, Laudet V. Retinoic acid influences anteroposterior positioning of epidermal sensory neurons and their gene expression in a developing chordate (amphioxus) Proc Natl Acad Sci U S A. 2004;101:10320–5. doi: 10.1073/pnas.0403216101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubert M, Yu J-K, Holland ND, Escriva H, Laudet V, Holland LZ. Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development. 2005;132:61–73. doi: 10.1242/dev.01554. [DOI] [PubMed] [Google Scholar]
- 42.Schubert M, Holland ND, Laudet V, Holland LZ. A retinoic acid-Hox hierarchy controls both anterior/posterior patterning and neuronal specification in the developing central nervous system of the cephalochordate amphioxus. Dev Biol. 2006;296:190–202. doi: 10.1016/j.ydbio.2006.04.457. [DOI] [PubMed] [Google Scholar]
- 43.Koop D, Chen J, Theodosiou M, Carvalho JE, Alvarez S, de Lera AR, et al. Roles of retinoic acid and Tbx1/10 in pharyngeal segmentation: amphioxus and the ancestral chordate condition. EvoDevo. 2014;5:36. doi: 10.1186/2041-9139-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koop D, Holland ND, Sémon M, Alvarez S, de Lera AR, Laudet V, et al. Retinoic acid signaling targets Hox genes during the amphioxus gastrula stage: insights into early anterior–posterior patterning of the chordate body plan. Dev Biol. 2010;338:98–106. doi: 10.1016/j.ydbio.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 45.Albalat R, Cañestro C. Identification of Aldh1a, Cyp26 and RAR orthologs in protostomes pushes back the retinoic acid genetic machinery in evolutionary time to the bilaterian ancestor. Chem Biol Interact. 2009;178:188–96. doi: 10.1016/j.cbi.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Bui-Göbbels K, Quintela RM, Bräunig P, Mey J. Is retinoic acid a signal for nerve regeneration in insects? Neural Regen Res. 2015;10:901–3. doi: 10.4103/1673-5374.158349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoppie P, Borgers M, Borghgraef P, Dillen L, Goossens J, Sanz G, et al. R115866 inhibits all-trans-retinoic acid metabolism and exerts retinoidal effects in rodents. J Pharmacol Exp Ther. 2000;293:304–12. [PubMed]
- 48.Thatcher JE, Buttrick B, Shaffer SA, Shimshoni JA, Goodlett DR, Nelson WL, et al. Substrate specificity and ligand interactions of CYP26A1, the human liver retinoic acid hydroxylase. Mol Pharmacol. 2011;80:228–39. doi: 10.1124/mol.111.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flood PR. Ciliary rootlet-fibres as tail fin-rays in larval amphioxus (Branchiostoma lanceolatum, Pallas). J Ultrastruct Res. 1975;51:218–25. [DOI] [PubMed]
- 50.Mansfield JH, Holland ND. Amphioxus tails: source and fate of larval fin rays and the metamorphic transition from an ectodermal to a predominantly mesodermal tail. Acta Zool. 2013;96:117–25. doi: 10.1111/azo.12058. [DOI] [Google Scholar]
- 51.Koop D, Holland LZ, Setiamarga D, Schubert M, Holland ND. Tail regression induced by elevated retinoic acid signaling in amphioxus larvae occurs by tissue remodeling, not cell death. Evol Dev. 2011;13:427–435. doi: 10.1111/j.1525-142X.2011.00501.x. [DOI] [PubMed] [Google Scholar]
- 52.Huang S, Chen Z, Yan X, Yu T, Huang G, Yan Q, et al. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat Commun. 2014;5:5896. doi: 10.1038/ncomms6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acemel RD, Tena JJ, Irastorza-Azcarate I, Marlétaz F, Gómez-Marín C, de la Calle-Mustienes E, et al. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat Genet. 2016;48:336–41. doi: 10.1038/ng.3497. [DOI] [PubMed] [Google Scholar]
- 54.Hu P, Tian M, Bao J, Xing G, Gu X, Gao X, et al. Retinoid regulation of the zebrafish cyp26a1 promoter. Dev Dyn. 2008;237:3798–808. [DOI] [PMC free article] [PubMed]
- 55.Zhang Y, Zolfaghari R, Ross AC. Multiple retinoic acid response elements cooperate to enhance the inducibility of CYP26A1 gene expression in liver. Gene. 2010;464:32–43. doi: 10.1016/j.gene.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pascual-Anaya J, Adachi N, Álvarez S, Kuratani S, D’Aniello S, Garcia-Fernàndez J. Broken colinearity of the amphioxus Hox cluster. EvoDevo. 2012;3:28. doi: 10.1186/2041-9139-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimozono S, Iimura T, Kitaguchi T, Higashijima S, Miyawaki A. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature. 2013;496:363–6. doi: 10.1038/nature12037. [DOI] [PubMed] [Google Scholar]
- 59.Holland LZ, Carvalho JE, Escriva H, Laudet V, Schubert M, Shimeld SM, et al. Evolution of bilaterian central nervous systems: a single origin? EvoDevo. 2013;4:27. doi: 10.1186/2041-9139-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bertrand S, Aldea D, Oulion S, Subirana L, de Lera AR, Somorjai I, et al. Evolution of the role of RA and FGF signals in the control of somitogenesis in chordates. PLoS ONE. 2015;10:e0136587. doi: 10.1371/journal.pone.0136587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schubert M, Holland LZ, Stokes MD, Holland ND. Three amphioxus Wnt genes (AmphiWnt3, AmphiWnt5, and AmphiWnt6) associated with the tail bud: the evolution of somitogenesis in chordates. Dev Biol. 2001;240:262–73. doi: 10.1006/dbio.2001.0460. [DOI] [PubMed] [Google Scholar]
- 62.Abu-Abed S, Dollé P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–40. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, Petkovich M. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birt Defects Res A Clin Mol Teratol. 2010;88:883–94. doi: 10.1002/bdra.20709. [DOI] [PubMed] [Google Scholar]
- 64.Wada H, Escriva H, Zhang S, Laudet V. Conserved RARE localization in amphioxus Hox clusters and implications for Hox code evolution in the vertebrate neural crest. Dev Dyn. 2006;235:1522–31. doi: 10.1002/dvdy.20730. [DOI] [PubMed] [Google Scholar]
- 65.Ahn Y, Mullan HE, Krumlauf R. Long-range regulation by shared retinoic acid response elements modulates dynamic expression of posterior Hoxb genes in CNS development. Dev Biol. 2014;388:134–44. doi: 10.1016/j.ydbio.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 66.Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Mol Cell Endocrinol. 2014;382:97–106. doi: 10.1016/j.mce.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 67.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–54. [PubMed] [Google Scholar]
- 68.Urushitani H, Katsu Y, Ohta Y, Shiraishi H, Iguchi T, Horiguchi T. Cloning and characterization of the retinoic acid receptor-like protein in the rock shell, Thais clavigera. Aquat Toxicol. 2013;142–143:403–13. [DOI] [PubMed]
- 69.Gutierrez-Mazariegos J, Nadendla EK, Lima D, Pierzchalski K, Jones JW, Kane M, et al. A mollusk retinoic acid receptor (RAR) ortholog sheds light on the evolution of ligand binding. Endocrinology. 2014;155:4275–86. doi: 10.1210/en.2014-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodríguez-Marí A, Cañestro C, BreMiller RA, Catchen JM, Yan Y-L, Postlethwait JH. Retinoic acid metabolic genes, meiosis, and gonadal sex differentiation in zebrafish. PLoS ONE. 2013;8:e73951. doi: 10.1371/journal.pone.0073951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- 73.Smith JJ, Keinath MC. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 2015;25:1081–90. doi: 10.1101/gr.184135.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Fritz A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 75.Barash MS. Mass extinction of the marine biota at the Ordovician-Silurian transition due to environmental changes. Oceanology. 2014;54:780–7. doi: 10.1134/S0001437014050014. [DOI] [Google Scholar]
- 76.Berner RA, VandenBrooks JM, Ward PD. Oxygen and evolution. Science. 2007;316:557–558. doi: 10.1126/science.1140273. [DOI] [PubMed] [Google Scholar]
- 77.Kremer B, Kaźmierczak J. Cyanobacterial mats from Silurian black radiolarian cherts: phototrophic life at the edge of darkness? J Sediment Res. 2005;75:897–906. doi: 10.2110/jsr.2005.069. [DOI] [Google Scholar]
- 78.Castle JW, Rodgers JH. Hypothesis for the role of toxin-producing algae in Phanerozoic mass extinctions based on evidence from the geologic record and modern environments. Environ Geosci. 2009;16:1–23. doi: 10.1306/eg.08110808003. [DOI] [Google Scholar]
- 79.Dudley R. Atmospheric oxygen, giant Paleozoic insects and the evolution of aerial locomotor performance. J Exp Biol. 1998;201:1043–50. doi: 10.1242/jeb.201.8.1043. [DOI] [PubMed] [Google Scholar]
- 80.Alroy J. The shifting balance of diversity among major marine animal groups. Science. 2010;329:1191–4. doi: 10.1126/science.1189910. [DOI] [PubMed] [Google Scholar]
- 81.Jia C, Huang J, Kershaw S, Luo G, Farabegoli E, Perri MC, et al. Microbial response to limited nutrients in shallow water immediately after the end-Permian mass extinction. Geobiology. 2012;10:60–71. doi: 10.1111/j.1472-4669.2011.00310.x. [DOI] [PubMed] [Google Scholar]
- 82.Berman-Frank I, Lundgren P, Falkowski P. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res Microbiol. 2003;154:157–64. doi: 10.1016/S0923-2508(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 83.Kelman D, Ben-Amotz A, Berman-Frank I. Carotenoids provide the major antioxidant defence in the globally significant N2-fixing marine cyanobacterium Trichodesmium. Environ Microbiol. 2009;11:1897–908. doi: 10.1111/j.1462-2920.2009.01913.x. [DOI] [PubMed] [Google Scholar]
- 84.Wu X, Jiang J, Wan Y, Giesy JP, Hu J. Cyanobacteria blooms produce teratogenic retinoic acids. Proc Natl Acad Sci. 2012;109:9477–82. doi: 10.1073/pnas.1200062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jonas A, Buranova V, Scholz S, Fetter E, Novakova K, Kohoutek J, et al. Retinoid-like activity and teratogenic effects of cyanobacterial exudates. Aquat Toxicol Amst Neth. 2014;155:283–90. doi: 10.1016/j.aquatox.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 86.Fuentes M, Schubert M, Dalfo D, Candiani S, Benito E, Gardenyes J, et al. Preliminary observations on the spawning conditions of the European amphioxus (Branchiostoma lanceolatum) in captivity. J Exp Zoolog B Mol Dev Evol. 2004;302B:384–391. doi: 10.1002/jez.b.20025. [DOI] [PubMed] [Google Scholar]
- 87.Fuentes M, Benito E, Bertrand S, Paris M, Mignardot A, Godoy L, et al. Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum) J Exp Zoolog B Mol Dev Evol. 2007;308B:484–493. doi: 10.1002/jez.b.21179. [DOI] [PubMed] [Google Scholar]
- 88.Theodosiou M, Colin A, Schulz J, Laudet V, Peyrieras N, Nicolas J-F, et al. Amphioxus spawning behavior in an artificial seawater facility. J Exp Zoolog B Mol Dev Evol. 2011;316B:263–275. doi: 10.1002/jez.b.21397. [DOI] [PubMed] [Google Scholar]
- 89.Holland LZ, Yu J-K. Cephalochordate (amphioxus) embryos: procurement, culture, and basic methods. Methods Cell Biol. 2004;74:195–215. doi: 10.1016/S0091-679X(04)74009-1. [DOI] [PubMed] [Google Scholar]
- 90.Yu JKS, Holland LZ. Extraction of RNA from amphioxus embryos or adult amphioxus tissue. Cold Spring Harb. Protoc. 2009;2009:pdb.prot5288. [DOI] [PubMed]
- 91.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 92.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 95.Huelsenbeck JP, Ronquist F. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 96.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2014.
- 97.Rambaut A. FigTree. 2012. Available from: http://tree.bio.ed.ac.uk/software/figtree/. Accessed 7 Oct 2016.
- 98.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Benton MJ, Donoghue PCJ, Asher RJ. Calibrating and constraining molecular clocks. The Timetree of Life. New York: Oxford University Press; 2009. p. 35–86.
- 100.dos Reis M, Thawornwattana Y, Angelis K, Telford MJ, Donoghue PCJ, Yang Z. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr Biol. 2015;25:2939–50. doi: 10.1016/j.cub.2015.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–23. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 103.Yu JKS, Holland LZ. Amphioxus whole-mount in situ hybridization. Cold Spring Harb. Protoc. 2009;2009:pdb.prot5286. [DOI] [PubMed]
- 104.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supporting data for this article are included in the supplementary information files. Additional raw data are available from the authors upon request.


