Abstract
Bloom syndrome is a rare disorder associated with cancer predisposition and genomic instability and is caused by loss of the RecQ helicase BLM. The Drosophila ortholog of BLM (DmBlm) is required for accurate repair of DNA double-strand gaps by homologous recombination. Repair products from DmBlm mutants have shorter repair synthesis tract lengths compared to wild type and are frequently associated with deletions flanking the break site. To determine the mechanisms responsible for deletion formation in the absence of DmBlm, we characterized repair after excision of the P{wa} element in various genetic backgrounds. Flies lacking DmRad51 do not have an elevated deletion frequency. Moreover, loss of DmRad51 suppresses deletion formation in DmBlm mutants. These data support a model in which DmBlm acts downstream of strand invasion to unwind a D-loop intermediate to free the newly synthesized strand. In the absence of DmBlm, alternative pathways of D-loop disassembly result in short repair synthesis tracts or flanking deletions. This model explains how RecQ helicases can promote homologous recombination while preventing illegitimate recombination.
DNA helicases of the RecQ family, which are conserved in organisms ranging from bacteria to mammals, play vital roles in the maintenance of genomic stability (reviewed in ref. 1). The importance of this family is highlighted by the observation that mutations in three of the five human RecQ helicases, WRN, BLM, and RECQ4, cause the Werner, Bloom, and Rothmund–Thomson syndromes, respectively (2–4). These three rare disorders are characterized by genomic instability and cancer predisposition, and patients with Werner and Rothmund–Thomson syndromes also display symptoms of premature aging.
Bloom syndrome (BS) patients develop a wide spectrum of cancers typical of those found in older individuals of the general population, including sarcomas, lymphomas, and epithelial cancers (reviewed in ref. 5). A hallmark of BS cells is a large increase in sister chromatid exchanges (6). In addition, BS cells show elevated levels of chromosome breaks, rearrangements, and deletions (7). Both in vivo and in vitro experiments with broken plasmids demonstrate that repair of double-strand breaks (DSBs) in the absence of BLM results in products with large deletions (8, 9). Therefore, a clearer understanding of the underlying events that cause chromosomal deletions in BS cells may provide insight into how the BLM protein prevents cancer in normal cells.
Accumulating evidence suggests that the BLM protein functions in homologous recombination repair pathways to promote genomic stability. In humans, BLM interacts with RAD51, a protein required for the strand invasion step that initiates DSB repair by homologous recombination (10). Sgs1p, the Saccharomyces cerevisiae homologue of BLM, also interacts with Rad51p (10), and sgs1 mutants have an increased rate of chromosomal rearrangements such as translocations and deletions. Interestingly, Sgs1p suppresses recombination between DNA sequences with imperfect homology (11), consistent with the notion that it functions both to promote accurate recombination and to suppress inappropriate recombination.
In vitro experiments provide further evidence that BLM functions during recombination. The human BLM helicase preferentially unwinds Holliday junctions, branched DNA structures, and other homologous recombination intermediates (12–14). Interestingly, BLM has also been shown to be adept at binding to and unwinding D-loops (15), which are thought to be the initial intermediate in DSB repair by homologous recombination.
The Drosophila melanogaster ortholog of BLM, DmBlm, is encoded by the mus309 gene (16). Mutations in mus309 cause increased sensitivity to ionizing radiation and defects in DSB repair. Reminiscent of the human phenotype, deletions flanking a DSB site on a plasmid are frequently observed in mus309 mutants (17, 18). We recently used a chromosomal DSB repair assay to demonstrate that mus309 mutants are defective in the repair of double-strand gaps created by the excision of a P transposable element (19).
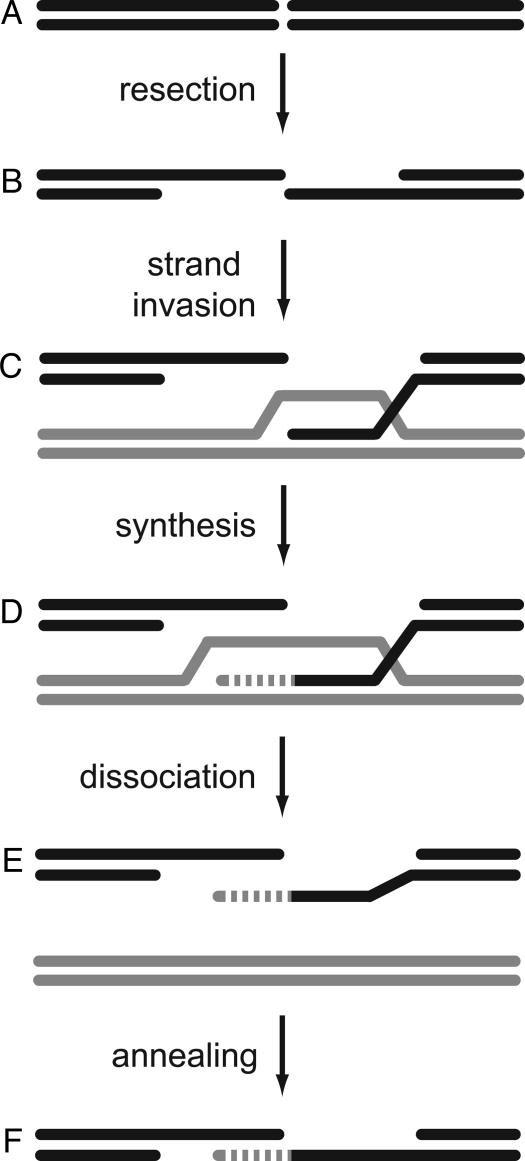
Double-strand gaps generated by P element excision are repaired predominantly through a homologous recombination pathway termed synthesis-dependent strand annealing (SDSA) (Fig. 1). During SDSA, single-stranded DNA is generated by 5′ to 3′ resection of each end. One or both of these single-stranded ends invade a homologous template and prime repair DNA synthesis. The newly synthesized strand is then dissociated from the template so that it can anneal to the single-stranded DNA from the other end of the break. Excision of a transposable element generates a gap relative to the sister chromatid, the preferred homologous template. We have proposed that repair DNA synthesis during SDSA is not highly processive, and that repair of a large gap involves repeated cycles of strand invasion, synthesis, and dissociation from one or both ends of the gap (20). If complementary sequences are found, repair can be completed to restore an intact element. Alternatively, the ends can be joined through a noncanonical end-joining pathway, even after multiple cycles of synthesis, resulting in an internally deleted element (20).
Fig. 1.
DSB repair by synthesis-dependent strand annealing. Processing of a DSB (A) begins with resection of the ends to leave 3′-ended single-stranded overhangs (B). One or both of these invade a homologous template (C) and prime repair synthesis (D). The nascent strand is dissociated (E), allowing it to anneal to complementary single-stranded DNA (F).
Several defects are apparent in P element excision repair products generated in the absence of DmBlm (19). As in the wild type, repair is usually initiated through the SDSA pathway. However, in the absence of DmBlm, SDSA rarely goes to completion. Instead, repair is completed by end joining. Synthesis tracts in the repair products are much shorter than in repair products generated in wild-type flies. In addition, many repair products are associated with deletions into sequences flanking the DSB site. These deletions may arise during an aberrant transposition process termed hybrid element insertion (21) or during aberrant repair processes. We have not observed any defects in transposition in mus309 mutants (unpublished data). Therefore, the highly elevated frequency of deletions in mus309 mutants is most likely the result of defects in repair caused by the absence of DmBlm.
Three nonexclusive models can explain the high incidence of deletion products in mus309 mutants. First, DmBlm or a complex that requires DmBlm may bind directly to the broken chromosome ends to prevent degradation by exonucleases. Second, DmBlm may be required for efficient repair synthesis during SDSA; in the absence of DmBlm, exonucleolytic activity predominates over synthesis, resulting in a net deletion of sequences. Third, DmBlm may act later during SDSA to promote the completion of repair DNA synthesis.
To test these models, we investigated the effect of mutating spn-A, which encodes DmRad51. In the absence of DmRad51, strand invasion cannot be initiated, preventing entry into SDSA. However, DSBs are still repaired efficiently through an end-joining pathway that utilizes small microhomologies within the 17-nt overhangs that remain after P element excision (20). We report here that these repair events are not usually associated with deletions, as is characteristic of repair events generated in the absence of DmBlm. Therefore, deletions are not simply a consequence of poor repair synthesis. We also show that removing DmRad51 suppresses the deletion phenotype of mus309 mutants. This result argues strongly that DmBlm is not required to protect DSB ends, and that it instead acts downstream of DmRad51 to prevent deletion formation. We propose a model that accounts for both the short repair synthesis tracts and the frequent deletions accompanying DSB repair in the absence of DmBlm.
Materials and Methods
Drosophila Stocks and Genetics. Flies were maintained on standard medium at 25°C. The P{wa} transgene used in this study is described by Kurkulos et al. (22) and Adams et al. (19). The mus309 mutants were compound heterozygotes of mus309D2 and mus309D3 (16). The spn-A mutants were compound heterozygotes of spnA057 and spnA093A (23). The transposase source used in these experiments was H{w+, Δ2–3}Hop2.1. Crosses for the P{wa} assay are described by Adams et al. (19) and McVey et al. (20). Briefly, single males containing P{wa} and transposase in a wild-type, mus309 mutant, or spn-A mutant background were crossed to four y w P{wa} virgins, and female progeny without transposase were scored. Aberrant repair products were recovered as white-eyed sons of yellow-eyed female progeny (only one female was taken from each cross vial). Absence of white-eyed sons indicated a male-lethal mutation. These were recovered instead in balanced daughters, and were confirmed to be sd deletions by crossing these to sd1 males and scoring the scalloped-wing phenotype of the daughters.
Molecular Analysis of Aberrant Repair. Repair synthesis tract lengths were determined as described in Adams et al. (19). Genomic DNA was prepared from single male flies containing the aberrant repair product. PCRs contained 10 mM Tris·HCl (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 1.25 μM each primer, 250 μM dNTPs, 2 μl of the genomic DNA prep, and TaqDNA polymerase in a 20-μl volume. PCR products were analyzed by agarose gel electrophoresis followed by ethidium bromide staining. Positive and negative controls were included in each set of reactions. Deletions flanking the DSB site were determined by a lack of PCR product using primers complementary to sequences of sd on both sides of the P{wa} insertion site (see Fig. 4). The nearest primers used were 200 bp to the left and 420 bp to the right.
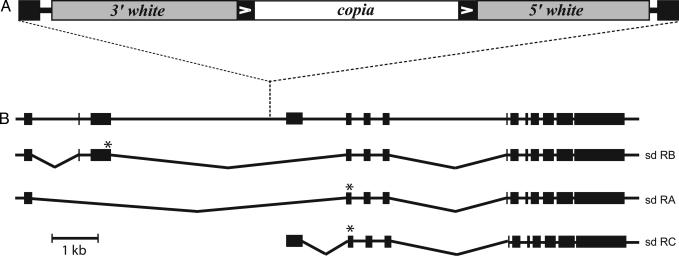
Fig. 4.
Schematic of P{wa} element inserted into sd.(A) P{wa} structure. Solid black rectangles indicate P ends. A copia retrotransposon (white) ending in LTRs (black rectangles with arrows) is inserted in the second intron of the white gene (gray). (B) Structure of sd. A map of sd exons (black boxes) is shown, with the three known transcripts depicted below. Coding start sites are marked with an asterisk; sd-RB begins at the end of the third exon, and sd-RA and -RC begin at the end of the fourth exon. The insertion site for P{wa} is indicated by the dashed line.
Results and Discussion
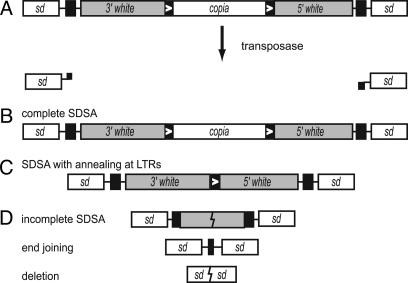
In the Absence of DmBlm, Most Repair Products Have Deletions Flanking the DSB Site. We previously established that the DmBlm protein is required for efficient repair of DNA DSBs created by excision of the P{wa} transposable element (19). In our experimental system, we generated males carrying an X-linked P{wa} insertion (Fig. 2) and a transposase source under control of a constitutive promoter. Transposase induces excision of P{wa} in both somatic and premeiotic germ-line tissues, leaving a DSB at the insertion site. Because P{wa} is located on the X chromosome, the only template for homologous repair in males is the sister chromatid; the DSB is actually a 14-kb gap relative to the sister chromatid. To recover individual repair events, these males are crossed to females homozygous for P{wa}, and repair events are initially scored according to the eye color of the female progeny. Most of these females have apricot-colored eyes, which indicates that the paternal P{wa} is intact. In most cases, this is probably because the element never was excised, but some of these may arise from excision followed by restoration of the entire P{wa} element by SDSA. SDSA can also result in red eyes (22); this class arises only through completed SDSA, and therefore provides an accurate indication of the relative efficiency of SDSA in various genetic backgrounds. Finally, yellow eye color indicates that the paternal P{wa} is absent or damaged, a class we refer to as “aberrant” repair. These events can arise from nonhomologous end joining, aborted SDSA (synthesis followed by end joining), or other mechanisms.
Fig. 2.
P{wa} excision creates DSBs that are repaired to produce distinct classes of products. (A) P{wa} inserted into an intron of scalloped (white, sd). P element inverted repeats (black) flank the white gene (gray), which contains a copia retrotransposon (white, copia) that has 276-bp LTRs (black with white arrows). Expression of transposase in males carrying P{wa} causes excision, resulting in a double-strand gap with 17-nt noncomplementary 3′ overhangs. The double-strand gap is then repaired through a variety of mechanisms. (B) Complete SDSA, resulting in restoration of P{wa}, gives apricot-colored eyes in females containing a maternally inherited copy of P{wa}. (C) SDSA with annealing at the LTRs produces a product with increased white expression, resulting in flat red eyes. (D) Aberrant repair includes aborted SDSA, end joining of the inverted repeats without synthesis, and deletions into flanking sequence. These events result in loss of white expression and are recovered as yellow-eyed progeny.
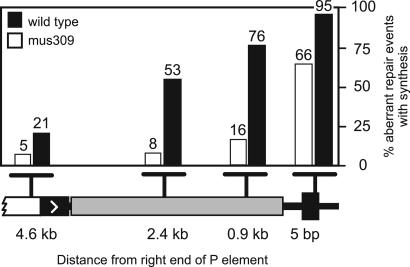
We previously reported that the red-eyed class was nearly absent among progeny of mus309 mutants, indicating a defect in SDSA (19). There was a corresponding increase in the yellow-eyed class, indicating aberrant repair in these mutants. Molecular and genetic analysis of these events revealed shorter synthesis tracts when DmBlm was absent, and frequent deletions into flanking sequences. The transposase source used to induce excision of P{wa} in these experiments is adjacent to the spn-A gene, so we are not able to generate spn-A mutants that carry this transposase source. Therefore, we repeated this experiment using a different transposase source, located on chromosome 2. Although this transposase appears to be more active in causing P element excision, it does not alter the way breaks are repaired (20). We found that the red-eyed class, which is indicative of completed SDSA, was reduced from 6% of progeny in the wild type (n = 3,624) to 0.2% in mus309 mutants (n = 2,851). In contrast, the yellow-eyed class, which is indicative of aberrant repair, was increased from 10% of progeny in the wild type to 15.5% in mus309 mutants. Aberrant repair products isolated from mus309 mutants had shorter repair DNA synthesis tract lengths compared to those isolated from the wild type (Fig. 3). These results are consistent with our previous observations using a different transposase source and confirm that the DmBlm protein is required for the extensive repair DNA synthesis of >14 kb needed to accurately repair the gap remaining after P{wa} excision.
Fig. 3.
Repair synthesis is reduced in mus309 mutants. Repair synthesis from the right end of the DSB was analyzed for 83 independent aberrant events in wild-type flies and 76 independent aberrant events in mus309 mutant flies. Molecular analysis was performed by using genomic DNA of males carrying a repair event from yellow-eyed mothers. The percentages of flies that had repair synthesis tract lengths at least as long as the indicated distance are shown.
The P{wa} transposon used in our assay is inserted into a 5-kb intron of the X-linked scalloped (sd) gene (Fig. 4), which encodes a transcription factor with homology to the human TEF-1 gene (24). Hypomorphic alleles, such as sd1, result in a scalloped-wing phenotype, but null alleles are lethal. We previously found that approximately one-quarter of the aberrant DSB repair events in mus309 males were accompanied by deletions that extended from the DSB site into a sd coding sequence, resulting in lethality in males and a scalloped-wing phenotype in females when in trans to sd1 (19). We observed similar results among the aberrant repair events generated in the experiment described above, in which a chromosome 2 transposase source was used: 2 of 85 (2.4%) from wild-type males had lethal sd deletions, whereas 26 of 104 (25.0%) from mus309 mutant males had lethal deletions (Table 1).
Table 1. Percentage of aberrant repair events with flanking deletions.
| Genotype | Lethal | Nonlethal | Total | n |
|---|---|---|---|---|
| Wild type | 2.4 | 7.0 | 9.4 | 85 |
| mus309 | 25.0 | 36.5 | 61.5 | 104 |
| spn-A | 2.6 | 1.7 | 4.3 | 116 |
| mus309 spn-A | 6.7 | 1.9 | 8.6 | 105* |
There were an additional four male-lethal chromosomes that did not produce a scalloped-wing phenotype in trans to sd1, indicating that these events did not involve deletions into sd. Similar events have been observed in other genotypes (M.M., unpublished data) and may result from transposition of the P{wa} into a nearby essential gene
Multiple transcripts of sd have been isolated (24), including one in which the first coding exon lies ≈3 kb upstream of the P{wa} insertion site, and two in which the first coding exon is <2 kb downstream of P{wa} (Fig. 4). We reasoned that some of our aberrant repair products could involve smaller deletions that were not male-lethal because they did not extend into sd coding sequence. To characterize these smaller deletions, we first determined whether there was a repair synthesis tract on each end of the DSB for each male-viable repair event. In cases where there was no repair synthesis tract on a given end of the DSB, we used PCR to determine whether there was a deletion from that end and, if so, to measure the extent of the deletion. Among 83 male-viable aberrant repair events from wild-type males, six had deletions of at least 200 bp to the left and two of these also had a deletion of at least 420 bp to the right. In total, 8 of the 85 aberrant repair events (9.4%) had deletions, two lethal and six nonlethal (Table 1). In contrast, among the 78 nonlethal aberrant repair events from mus309 mutants, 33 had deletions to the left; five of these also had deletions to the right, and five additional nonlethal repair events had a deletion only to the right. In total, of 104 aberrant repair events from mus309 mutants, 64 (61.5%) had deletions (Table 1).
In all instances, we recovered more nonlethal deletions to the left of P{wa} than to the right. This apparent bias may be a consequence of the P{wa} insertion site rather than a difference in repair from the left versus the right side of the break. The presence of essential sd coding information <2 kb to the right of P{wa} likely results in the recovery of many right-side deletions as male-lethal events; thus, we recover a smaller relative percentage of nonlethal deletions to the right of P{wa}.
Accounting for the percentage of progeny that had aberrant repair events, 0.94% of total progeny from wild-type males had deletions, but 9.5% of total progeny from mus309 mutants had deletions, a 10-fold increase. We have estimated that only 20% of all progeny are derived from cells that have experienced an excision event (20). When we used this estimate, we detected deletions of at least 200 bp in <5% of all repair events generated in wild-type males, but in 47% of repair events generated in the absence of DmBlm. Because cells from BS patients frequently have chromosomal deletions (7), understanding the mus309 deletion phenotype may provide clues about the molecular function of the BLM protein. Therefore, we undertook a more extensive genetic and molecular analysis to determine potential mechanisms of deletion formation.
Flanking Deletions Are Not Caused by a Lack of Repair Synthesis During SDSA. Repair synthesis tracts from mus309 aberrant repair events are significantly shorter than those from wild-type flies, suggesting a potential defect in repair synthesis (19) (Fig. 3). One possible explanation for the large number of deletions in mus309 mutants is that they are a consequence of a block to efficient repair DNA synthesis. This model predicts that, if repair synthesis is completely blocked, the deletion phenotype should be at least as severe as that observed in mus309 mutants.
Repair synthesis requires invasion of the broken DNA strand into a homologous template. The Rad51 protein is known to catalyze this strand exchange reaction in multiple organisms (reviewed in refs. 25 and 26). In flies, the DmRad51 protein is encoded by the spn-A gene (23). Flies lacking DmRad51 are unable to carry out DNA synthesis during DSB repair, and instead join the broken ends together without synthesis (20). To determine whether a synthesis defect is required for deletions, we used the P{wa} assay to analyze repair events in spn-A mutants.
We characterized 116 aberrant repair events isolated from spn-A mutant males, and found that only three (2.6%) were male-lethal, indicating deletions into sd coding sequence (Table 1). An additional two (1.7%) had nonlethal deletions, and these were similar in size to those isolated from the wild type and mus309 mutants. Thus, a total of 4.3% of aberrant repair events in flies lacking DmRad51 had flanking deletions; this frequency is not significantly different from the frequency in the wild type (P = 0.16). Because spn-A mutants have greatly reduced repair synthesis relative to mus309 flies, but mus309 repair products are 14 times more likely to involve deletions, it follows that a lack of repair synthesis per se does not lead to deletion formation.
Flanking Deletions Occur After Strand Invasion. The flanking deletions associated with DSB repair in the absence of DmBlm do not appear to be a result of decreased repair synthesis. An alternative model is that DmBlm is involved in protecting the broken DNA ends from degradation, either by binding to the ends or by recruiting a complex that binds to the ends. If this model is correct, then mus309 spn-A double mutants should still have a high rate of deletions because they lack DmBlm to protect the ends.
Therefore, we repeated the P{wa} assay in mus309 spn-A double mutant males. As in the spn-A single mutants, we did not recover any red-eyed progeny (n = 3,606); 17% had yellow eyes. We molecularly analyzed 109 independent repair products from yellow-eyed progeny. Seven (6.4%) were male lethal, indicating deletions into sd coding sequences (Table 1). An additional two repair events were associated with nonlethal flanking deletions, for a total of 8.6%. The frequency of deletions in the mus309 spn-A double mutants is not significantly different from the frequency in the wild type (P = 0.8). We conclude that the large increase in deletions generated in the absence of DmBlm occurs only when DmRad51 is present.
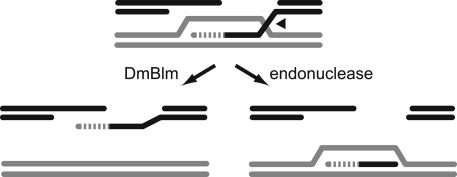
Model for DSB Repair in Wild-Type and mus309 Mutant Flies. We have reported that DSB repair in the absence of DmBlm is associated with frequent deletions into flanking sequences. Removing DmRad51 suppresses this deletion phenotype, suggesting that DmBlm functions after DmRad51 in preventing deletion formation. One possibility is that DmBlm acts before the repair synthesis fork to unwind the template. This could account for the shorter synthesis tracts found in repair events that take place in the absence of DmBlm. However, it is difficult to see how defects in this function could result in deletions. A more attractive alternative is that DmBlm acts behind the synthesis fork to unwind the nascent strand (Fig. 5). In SDSA, the ends of a break are resected to generate 3′ single-stranded tails. One (or both) of these invades a homologous template to form a D-loop, and the tail is then extended by repair DNA synthesis. The hallmark of SDSA is the annealing of this nascent strand to the single-stranded DNA at the other end of the break. For this to occur, the nascent strand must first be dissociated from the template, resolving the D-loop. We propose that DmBlm is involved in this dissociation step.
Fig. 5.
Model for the function of DmBlm in DSB repair. (Upper) The intermediate after strand invasion and repair synthesis. DmBlm functions at the junction marked by the arrowhead to unwind the invading strand. The resulting single-stranded DNA can then anneal to a complementary strand (Lower Left). In the absence of DmBlm, an endonuclease cleaves the invading strand at the arrowhead. The result (Lower Right) is a double-stranded deletion of sequences adjacent to the original DSB site.
Dissociation of the nascent strand could cause termination of repair synthesis, resulting in a lack of processivity. If DmBlm is responsible for dissociation of the nascent strand, then we might expect to see longer synthesis tracts in the absence of DmBlm. However, we recover shorter synthesis tracts in the absence of DmBlm function, suggesting that DmBlm is not responsible for the lack of processivity during repair synthesis.
In the absence of dissociation by DmBlm, the D-loop intermediate must be processed by an alternative mechanism. We propose that one way to separate the two duplexes is to cut the invading strand 5′ to the point of strand transfer (Fig. 5 Lower Right). According to this proposal, after DSB formation, one strand is resected from the end of the break. The single-stranded region exposed by resection invades a homologous template, and subsequently is cleaved. This combination of resection of one strand and cleavage of the complementary strand results in a double-stranded deletion of sequences flanking the original break point.
In conditions where end joining is unfavorable, this deletion could be enlarged through multiple cycles of reresection, invasion, and cleavage. In a previous study, we suggested that repair synthesis is not highly processive, and estimated that the median tract length may be only a few hundred nucleotides (20). Although this would be sufficient for repair of a simple DSB, it is unlikely that a single episode of repair synthesis would traverse the 14-kb gap remaining after excision of the P{wa} element. Therefore, SDSA repair of a large gap must require multiple cycles of strand invasion, synthesis, and dissociation, perhaps occurring independently at both ends. In the absence of DmBlm, repair by end joining after many rounds of resection, invasion, and cleavage could generate the large deletions that we observe.
Although most repair events isolated from mus309 mutants involved deletions, many did have detectable synthesis tracts. Therefore, it must be possible to dissociate the nascent strand in some cases. One possible explanation is that the mus309 mutants we used were not completely devoid of DmBlm activity. The males used in our assay are derived from mothers that are heterozygous for a mus309 mutation, so there may have been perdurance of some maternally derived DmBlm. We have observed a strong maternal effect for heterozygosity for spn-A (20). Alternatively, DmBlm may not be absolutely required to unwind the D-loop. There are two other RecQ helicases in Drosophila, RecQ5 and RecQ4 (27), and it is possible that one of these or some other helicase can weakly compensate for loss of DmBlm. In either scenario, a model in which multiple rounds of invasion, synthesis, and dissociation are required to repair across a large gap predicts that in mus309 mutants, dissociation will ultimately fail. Alternative processing of the D-loop, followed by end joining, will result in the net decrease in synthesis tract lengths that we observe.
D-loops such as those found during SDSA repair synthesis are a preferred in vitro substrate for human BLM and E. coli RecQ (15, 28), lending support to our model. BLM interacts physically with RAD51 (10). Because RAD51 coats the single-stranded DNA before D-loop formation (29), this interaction might provide a mechanism for targeting BLM to its preferred substrate in SDSA.
Our model provides an explanation for the paradoxical finding that RecQ helicases can both prevent illegitimate recombination and promote homologous recombination (11, 28, 30, 31). Unwinding of D-loops generated during strand invasion into a nonhomologous or homeologous template can prevent illegitimate recombination, whereas unwinding of D-loops after repair synthesis using a homologous template can promote homologous recombination.
The identity of the endonuclease we propose is unknown. Mutation of the S. cerevisiae BLM ortholog, SGS1, is synthetically lethal with mutations in any of six SLX genes, four of which encode the MUS81/MMS4 and SLX1/SLX4 heterodimeric structure-specific endonucleases (32). Interestingly, mutations in RAD51 rescue the synthetic lethality between sgs1 and mus81 or mms4 (33). Orthologs of MUS81, MMS4, and SLX1 can be identified in Drosophila (unpublished data). Given the genetic interactions in S. cerevisiae, it will be interesting to determine whether any of these genes plays a role in deletion formation after aberrant DSB repair in flies lacking DmBlm.
In summary, we have demonstrated that loss of DmBlm results in a propensity to repair DSBs by a mechanism that produces large deletions. Because the formation of these deletions requires Rad51-mediated strand invasion, the primary function of DmBlm in SDSA must occur after strand invasion. We propose that DmBlm is required to dissociate the invading strand, and the synthesis tract primed off it, from the homologous template to allow annealing to a complementary single strand. In the absence of DmBlm, this D-loop structure is cleaved, generating a deletion. The identification of an endonuclease that promotes disassembly of D-loops in the absence of DmBlm could provide insight into error-prone repair mechanisms that operate in BS, causing genomic instability and cancer.
Acknowledgments
We thank Heather Yarnall-Schultz, Hunter Blanton, and Sarah Radford for helpful comments on the manuscript. J.R.L. was supported by aGraduate Research Fellowship from the National Science Foundation. M.M. was supported by Minority Opportunities in Research Division of the National Institute of General Medical Sciences (NIGMS) Grant GM-000678. This work was supported by a New Scholar Award from the Ellison Medical Foundation and NIGMS Grant R01 GM-61252 (to J.J.S.).
Author contributions: M.M., J.R.L., M.D.A., and J.J.S. designed research; M.M., J.R.L., and M.D.A. performed research; M.M., J.R.L., M.D.A. and J.J.S. analyzed data; and M.M., J.R.L., and J.J.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BS, Bloom syndrome; DSB, double-strand break; SDSA, synthesis-dependent strand annealing.
References
- 1.Hickson, I. D. (2003) Nat. Rev. Cancer 3, 169-178. [DOI] [PubMed] [Google Scholar]
- 2.Yu, C.-E., Oshima, J., Fu, Y.-H., Wijsman, E. M., Hisama, F., Alisch, R., Metthews, S., Nakura, J., Miki, T., Ouais, S., et al. (1996) Science 272, 258-262. [DOI] [PubMed] [Google Scholar]
- 3.Ellis, N. A., Groden, J., Ye, T.-Y., Straughen, J., Lennon, D. J., Ciocci, S., Proytcheva, M. & German, J. (1995) Cell 83, 655-666. [DOI] [PubMed] [Google Scholar]
- 4.Kitao, S., Shimamoto, A., Goto, M., Miller, R. W., Smithson, W. A., Lindor, N. M. & Furuichi, Y. (1999) Nat. Genet. 22, 82-84. [DOI] [PubMed] [Google Scholar]
- 5.German, J. (1993) Medicine 72, 393-406. [PubMed] [Google Scholar]
- 6.Chaganti, R. S., Schonberg, S. & German, J. (1974) Proc. Natl. Acad. Sci. USA 71, 4508-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tachibana, A., Tatsumi, K., Masui, T. & Kato, T. (1996) Mol. Carcinog. 17, 41-47. [DOI] [PubMed] [Google Scholar]
- 8.Runger, T. M. & Kraemer, K. H. (1989) EMBO J. 8, 1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaymes, T. J., North, P. S., Brady, N., Hickson, I. D., Mufti, G. J. & Rassool, F. V. (2002) Oncogene 21, 2525-2533. [DOI] [PubMed] [Google Scholar]
- 10.Wu, L., Davies, S. L., Levitt, N. C. & Hickson, I. D. (2001) J. Biol. Chem. 276, 19375-19381. [DOI] [PubMed] [Google Scholar]
- 11.Myung, K., Datta, A., Chen, C. & Kolodner, R. D. (2001) Nat. Genet. 27, 113-116. [DOI] [PubMed] [Google Scholar]
- 12.Karow, J. K., Chakraverty, R. K. & Hickson, I. D. (1997) J. Biol. Chem. 272, 30611-30614. [DOI] [PubMed] [Google Scholar]
- 13.Karow, J. K., Constantinou, A., Li, J. L., West, S. C. & Hickson, I. D. (2000) Proc. Natl. Acad. Sci. USA 97, 6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohaghegh, P., Karow, J. K., Brosh, R. M., Jr., Bohr, V. A. & Hickson, I. D. (2001) Nucleic Acids Res. 29, 2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Brabant, A. J., Ye, T., Sanz, M., German, I. J., Ellis, N. A. & Holloman, W. K. (2000) Biochemistry 39, 14617-14625. [DOI] [PubMed] [Google Scholar]
- 16.Kusano, K., Johnson-Schlitz, D. M. & Engels, W. R. (2001) Science 291, 2600-2602. [DOI] [PubMed] [Google Scholar]
- 17.Beall, E. L. & Rio, D. C. (1996) Genes Dev. 10, 921-933. [DOI] [PubMed] [Google Scholar]
- 18.Min, B., Weinert, B. T. & Rio, D. C. (2004) Proc. Natl. Acad. Sci. USA 101, 8906-8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams, M. D., McVey, M. & Sekelsky, J. J. (2003) Science 299, 265-267. [DOI] [PubMed] [Google Scholar]
- 20.McVey, M., Adams, M. D., Staeva-Vieira, E. & Sekelsky, J. J. (2004) Genetics 167, 699-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preston, C. R. & Engels, W. R. (1996) Genetics 144, 1623-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurkulos, M., Weinberg, J. M., Roy, D. & Mount, S. M. (1994) Genetics 136, 1001-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staeva-Vieira, E., Yoo, S. & Lehmann, R. (2003) EMBO J. 22, 5863-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell, S., Inamdar, M., Rodrigues, V., Raghavan, V., Palazzolo, M. & Chovnick, A. (1992) Genes Dev. 6, 367-379. [DOI] [PubMed] [Google Scholar]
- 25.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung, P., Krejci, L., Van Komen, S. & Sehorn, M. G. (2003) J. Biol. Chem. 278, 42729-42732. [DOI] [PubMed] [Google Scholar]
- 27.Sekelsky, J. J., Brodsky, M. H. & Burtis, K. C. (2000) J. Cell Biol. 150, F31-F36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harmon, F. G. & Kowalczykowski, S. C. (1998) Genes Dev. 12, 1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumann, P. & West, S. C. (1997) EMBO J. 16, 5198-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanada, K., Ukita, T., Kohno, Y., Saito, K., Kato, J.-I. & Ikeda, H. (1997) Proc. Natl. Acad. Sci. USA 94, 3860-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamagata, K., Kato, J., Shimamoto, A., Goto, M. & Furuichi, Y. (1998) Proc. Natl. Acad. Sci. USA 95, 8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullen, J. R., Kaliraman, V., Ibrahim, S. S. & Brill, S. J. (2001) Genetics 157. [DOI] [PMC free article] [PubMed]
- 33.Fabre, F., Chan, A., Heyer, W. D. & Gangloff, S. (2002) Proc. Natl. Acad. Sci. USA 99, 16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]