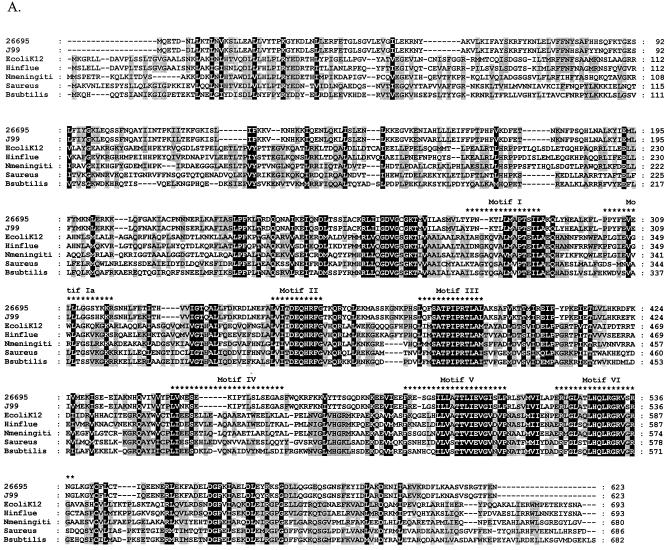
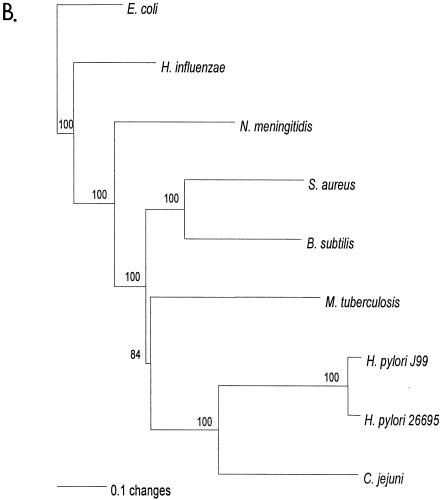
FIG. 1.
Relationship of deduced H. pylori products with RecG homologs. A. Alignment of amino acid sequences of H. pylori HP1523, JHP1412, and RecG homologs from five bacterial species (E. coli K-12, Haemophilus influenzae [Hinflue], Neisseria meningitidis [Nmeningiti], S. aureus, and Bacillus subtilis). Protein sequences were aligned with ClustalW, and the locations of conserved RecG helicase motifs (35) are indicated with asterisks. The program Genedoc (www.psc.edu/biomed/genedoc) was used to visualize the alignment in quantify mode, which highlights the one, two, or three most-frequent residues found in each column of the alignment. Conservative substitutions (e.g., I, L, V, M) were treated as if identical. Amino acids conserved in all species are indicated by white letters on black background. Gaps introduced to maximize alignment are indicated by dashes. Black letters on gray shading represent 60% or greater identity. White letters on black shading represent 100% identity. B. Phylogeny of RecG orthologs. Phylogenetic analysis of RecG orthologs was performed on 8 representative bacterial species, with bootstrap values listed next to the branches. M. tuberculosis, Mycobacterium tuberculosis.