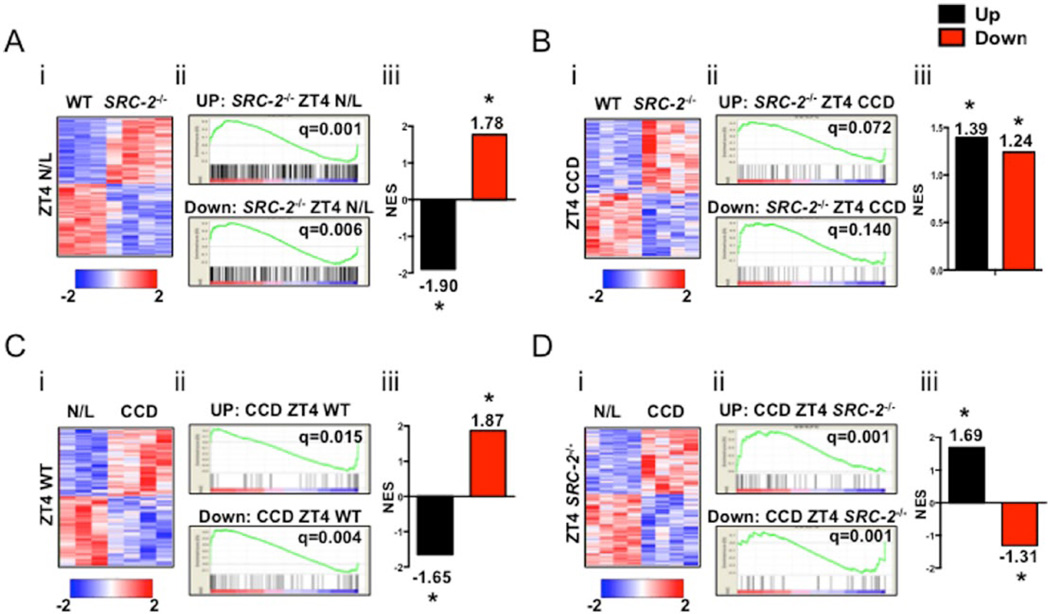
Figure 4.
Gene set enrichment analysis (GSEA) of murine CCD hepatic transcriptomic data compared with human HCC microarray data. (A) (i) Heat map representation of liver microarray in WT and SRC-2−/− mice at ZT4 N/L. (ii) GSEA plot representation of upregulated and down-regulated genes in common between the HCC human hepatic microarray dataset and enriched relative to WT in the SRC-2−/− ZT4 N/L hepatic microarray. (iii) Normalized enrichment score of up-regulated and down-regulated genes from A(ii). (B) (i) Heat map representation of liver microarray in WT and SRC-2−/− mice at ZT4 CCD. (ii) GSEA plot representation of up-regulated and down-regulated genes in common between the HCC human hepatic microarray dataset and enriched relative to WT in the SRC-2−/− ZT4 CCD hepatic microarray. (iii) Normalized enrichment score of up-regulated and down-regulated genes from B(ii). (C) (i) Heat map representation of liver microarray in N/L and CCD WT mice at ZT4. (ii) GSEA plot representation of up-regulated and down-regulated genes in common between the HCC human hepatic microarray dataset and enriched relative to N/L in the CCD ZT4 WT mice hepatic microarray. (iii) Normalized enrichment score of up-regulated and down-regulated genes from C(ii). (D) (i) Heat map representation of liver microarray in N/L and CCD SRC-2−/− mice at ZT4. (ii) GSEA plot representation of up-regulated and down-regulated genes in common between the HCC human hepatic microarray dataset and enriched relative to N/L in the CCD ZT4 SRC-2−/− mice hepatic microarray. (iii) Normalized enrichment score of up-regulated and down-regulated genes from D(ii).