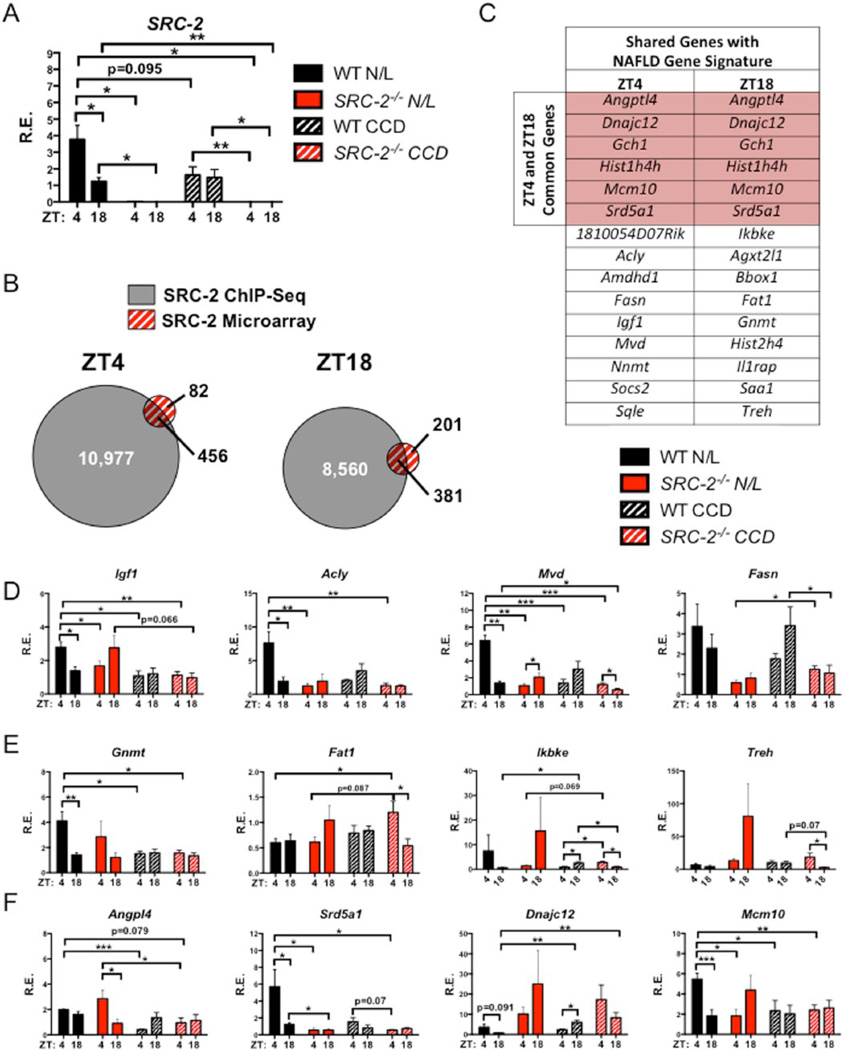
Figure 5.
Hepatic transcriptomic analysis of WT and SRC-2−/− mice subjected to N/L and CCD. (A) Liver mRNA expression of SRC-2 in WT and SRC-2−/− mice subjected to N/L and CCD at ZT4 and ZT18. (B) Overlap of hepatic ZT4 (left) and ZT18 (right) SRC-2 ChIP-Seq and microarray data from SRC-2−/− mice maintained on N/L at ZT4 and ZT18. (C) Table of common genes shared between the SRC-2 ChIP-Seq and microarray from ZT4 and ZT18 from (B) that are found in common with human liver microarrays from patients with NASH and HCC. (D) mRNA expression of genes from (C) that represent ZT4 genes that are found in common with human patients with NASH and HCC in WT and SRC-2−/− mice subjected to N/L and CCD at ZT4 and ZT18. (E) mRNA expression of genes from (C) that represent ZT18 genes that are found in common with human patients with NASH and HCC in WT and SRC-2−/− mice maintained on N/L and CCD conditions at ZT4 and ZT18. (F) mRNA expression of genes from (C) that represent ZT4 and ZT18 genes that are found in common with human patients with NASH and HCC in WT and SRC-2−/− mice subjected to N/L and CCD at ZT4 and ZT18. Data are graphed as the mean ± SEM. *p < 0.05. **p < 0.01. ***p < 0.001. (N = 3–4).