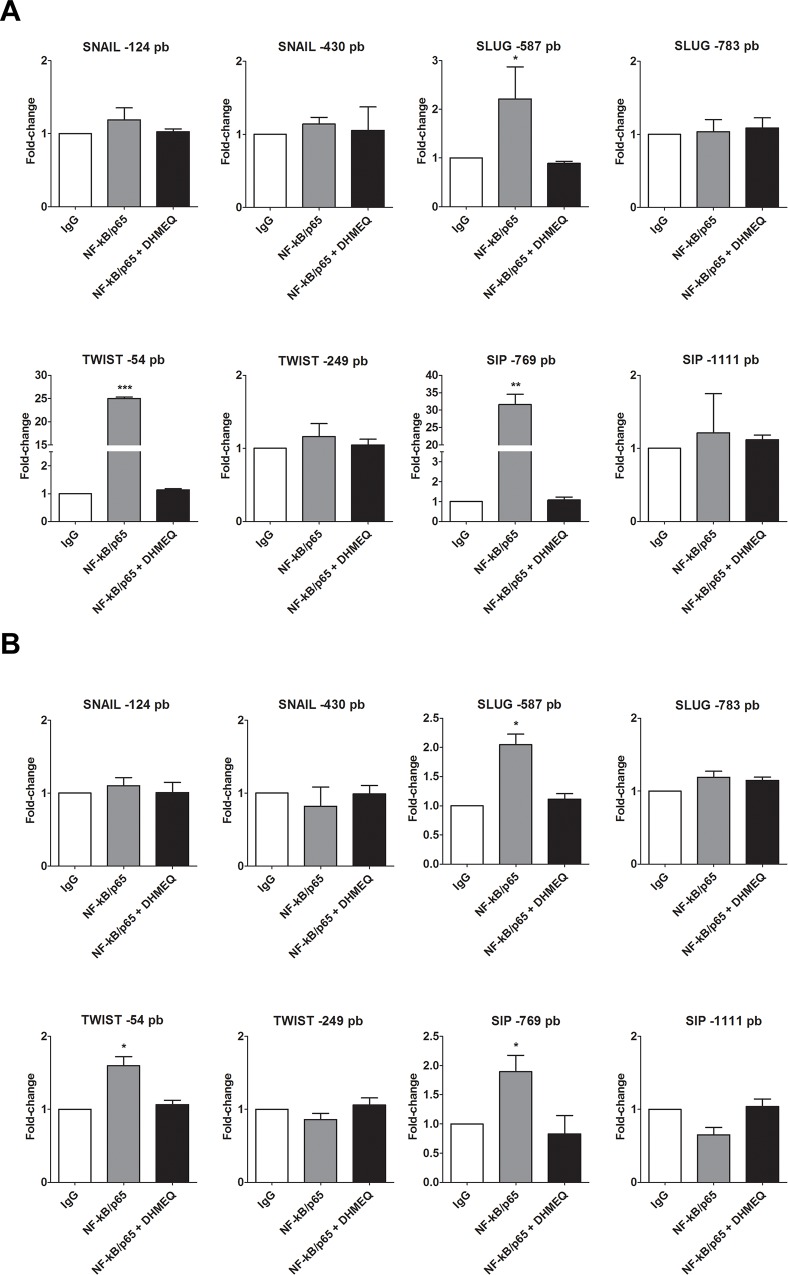
Fig 7.
ChIP-qPCR of predicted NF-κB/p65 binding sites in the SNAIL1, SLUG, TWIST1 and SIP1 promoter regions using MDA-MB-231 (A) and HCC-1954 (B) cells. The histograms set a fold-change of each site by comparing the IgG negative control to NF-κB/p65 antibodies with the natural and treated (DHMEQ) condition. The data were expressed as the mean ± SD. * = p<0.05, ** = p<0.01, *** = p<0.001.