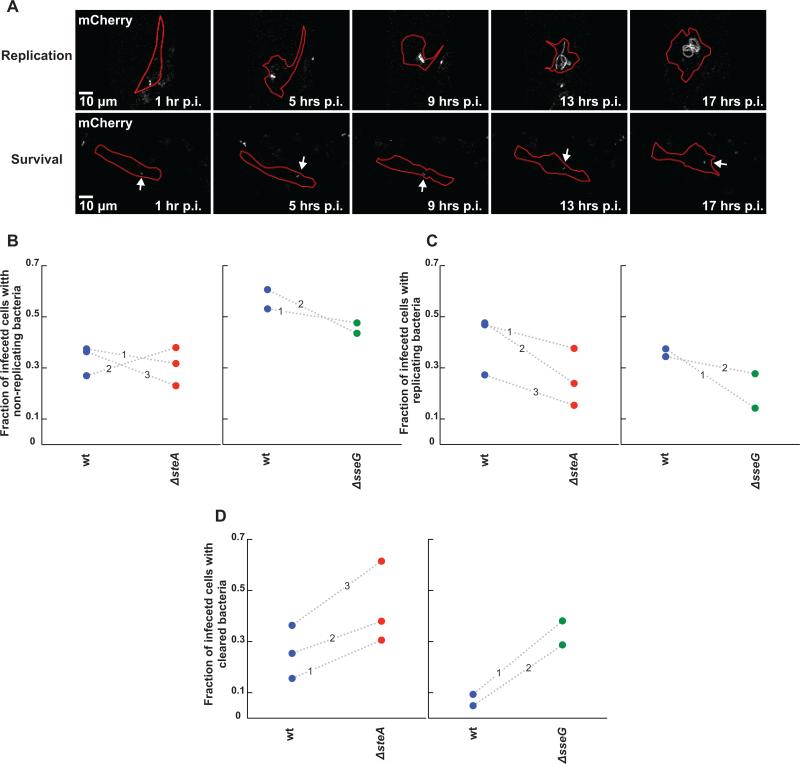
Figure 5.
Assay for monitoring the roles of SteA and SseG in Salmonella persistence, replication, or clearance in single primary bone marrow derived macrophages (BMDMs). (A) Representative fluorescence images of mCherry Salmonella replication (top panel) or survival (non-replicating state, bottom panel, labeled with an arrow) at distinct time points post infection. Salmonella is in gray and cell outlines are marked in red. (B-D) Comparison of ΔsteA (red circles) or ΔsseG (green circles) to wt Salmonella (blue circles) in altering the fraction of infected cells containing persisting (B), replicating (C), or cleared bacteria (D) between 1 and 16 hours post infection. Shown here is the fraction of cells exhibiting each phenotype for each strain. Each circle represents a fraction from a single independent experiment. Wt and mutant fractions from the same experiment are connected by a dotted line marked by the experiment number. (B) Deletion of either steA or sseG does not alter the fraction of cells containing persisting bacteria compared to wt Salmonella. (C) Deletion of both steA and sseG decrease the fraction of cells containing replicating bacteria. (D) ΔsteA or ΔsseG bacteria are more easily cleared by BMDMs compared to wt bacteria, though deletion of sseG causes a much stronger phenotype.