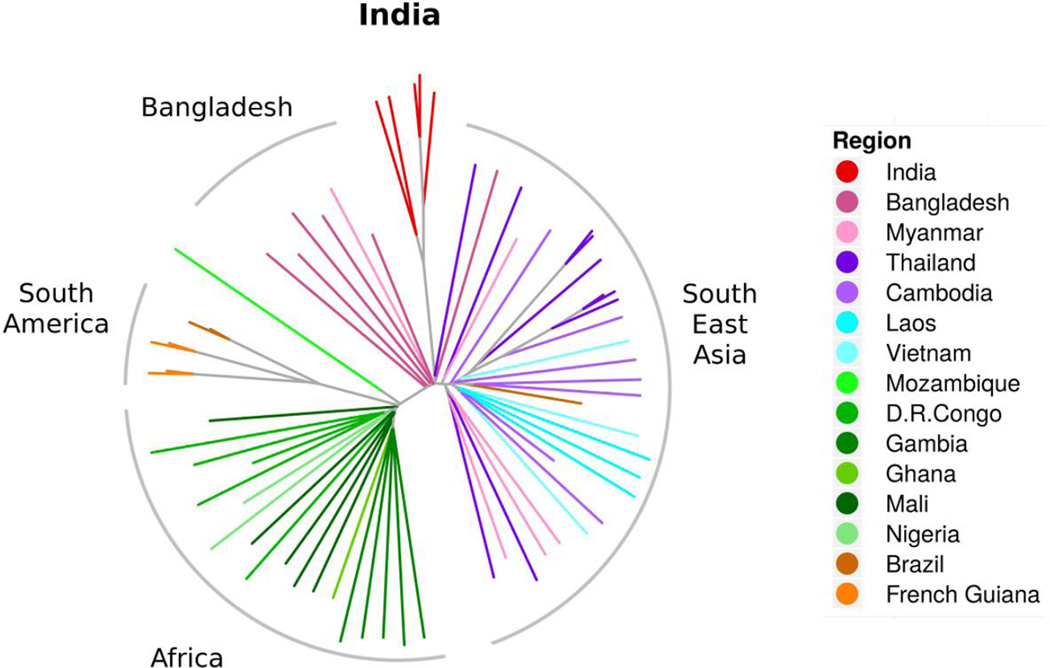
Figure 2.
Positioning of P. falciparum genomes from India amongst global isolates of the human malaria parasites using un-rooted neighbor joining tree, excluding var genes and non-coding sequences. Non-coding sequences and variable surface protein genes were removed and the distance analysis was based on coding sequences where basic cellular metabolism and physiology functions must reside. This grouping reflected true geographic origins of the parasite samples. African samples showed preferential joining of parasites from Congo and from Gambia. Parasites from West and Central Africa were well-separated from the Mozambique parasite. In Southeast Asia, there was a continual separation of parasites from Bangladesh to Myanmar/Thailand, to Cambodia, and to Laos and Vietnam. Parasites in Thailand were less tightly concentrated, but may reflect the movement of patients and parasites in and out of this country. In this context, the Indian P. falciparum isolates remained cleanly segregated from Southeast Asian, African, South American and Bangladesh lines. The continental clusters were well-defined with minimum ambiguity.
In an earlier effort, we performed distance analysis using whole genomes (including var genes and intergenic regions) superimposed on the P. falciparum 3D7 cell line. This generated some uncertain relationships between isolates and their geographic origins (data not shown). We hypothesize that the ancestral genomic relationships of the parasites may be influenced by subsets of highly-variable parasite genes formed based on interactions with patient immune responses. Genes coding for surface proteins on infected erythrocytes are under heavy selective pressure and reshuffle amongst themselves at very high rates and acquire point mutations [22, 23]. In addition, non-coding DNA spread across the chromosome forms a substantial part of the parasite genome and could also drift more compared to internal protein coding genes. As shown above, removal of var genes and intergenic regions provided a reliable picture of relationships between genomes and origin of samples.