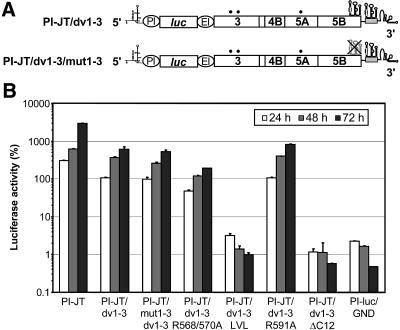
FIG. 6.
Discrimination of protein versus RNA effects of mutations in the NS5B insertion sequence. (A) Schematic structures of replicons with a duplication of SL3 in the variable region of the 3′ NTR. The gray box indicates the duplication of part of the 3′ coding region of NS5B that carries the predicted stem-loop structures. Replicon PI-JT/dv1-3 contained two identical intact copies of the SL3 sequence and was used as a parental construct to introduce the NS5B insertion sequence mutations. The predicted SL3 RNA secondary structure in the coding region of NS5B was disrupted by multiple silent mutations in replicon PI-JT/mut1-3/dv1-3. For further details, see the legend to Fig. 5A. (B) Replicons containing two intact copies of SL3 and the indicated mutations in the NS5B insertion sequence were transfected into Huh-7/lunet cells by electroporation. The percentage of luciferase activity at a given time point is relative to the relative light units determined 4 h after transfection, which was set to 100%. Replicon PI-luc/GND, which contains an inactivating mutation in the GDD motif of NS5B, was used as a negative control.