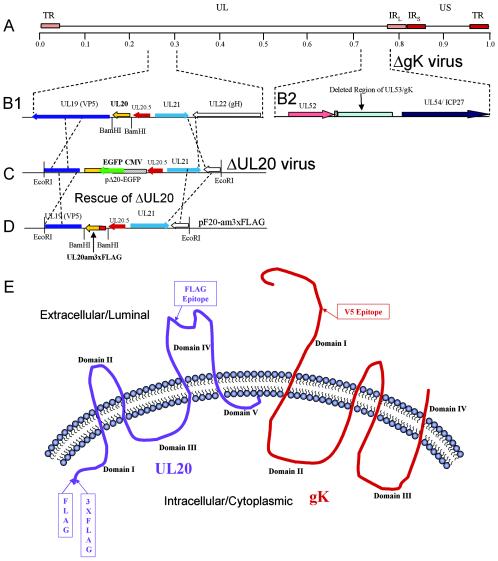
FIG. 1.
Schematic of gK-null and UL20-null recombinant viruses expressing epitope-tagged UL20p and gK, respectively. (A) The top line represents the prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (B) An expanded genomic region between map units 0.25 and 0.3 containing the UL19, UL20, UL20.5, UL21, and UL22 genes (panel 1) or the region between map units 0.7 and 0.8 containing the UL52, UL53, and UL54 open reading frames (panel 2). (C) The recombination plasmid, pΔ20-EGFP, which contains UL20 flanking sequences for recombination with the viral genome, was used through homologous recombination with the ΔgK viral genome to generate a ΔUL20/ΔgK double-null virus. (D) The recombination plasmid pF20-am3xFLAG, which encodes a 3xFLAG epitope-tagged UL20 gene and flanking sequences for recombination, was used to rescue the UL20 deletion and transfer the 3xFLAG epitope into the virus. (E) Schematic depicting the experimentally determined membrane topology of gK and the predicted membrane topology of UL20p, as well as the relative sites of insertion of each of the epitope tags.