FIG. 5.
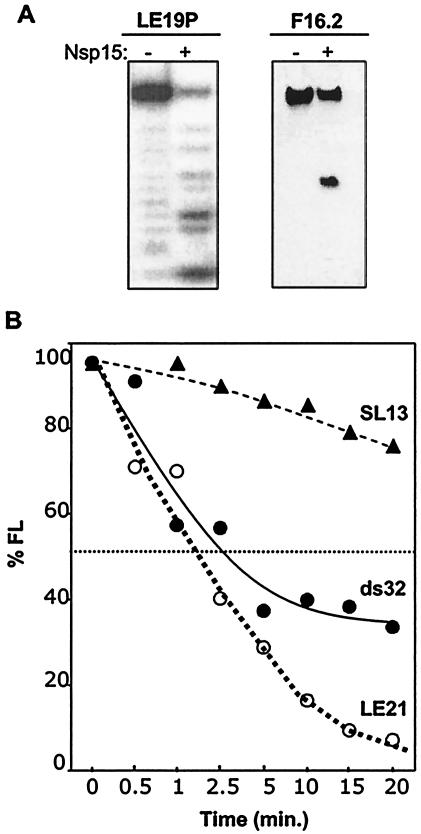
Substrate requirements for endoribonucleolytic activity of SARS-CoV Nsp15. (A) RNA substrates with chemically modified 3′ and 5′ termini. RNA LE19P, with a covalently attached 3′ puromycin is a weakly base-paired RNA that could form a weakly stable intramolecular hairpin or partially base-paired dimer (19). F16.2 is a 16-nt RNA that was synthesized with a 5′ fluorescein. The amount of cleavage was normalized to the percentage of full-length RNA remaining after the treatment (% FL). The RNA used in each experiment and whether Nsp15 was added (+) or not (−) are indicated above the gels. Digestion of LE19P is shown as a PhosphorImage of radiolabeled RNA, while F16.2 was from a toluidine blue-stained gel. (B) Comparison of the rate of cleavage of single- and double-stranded RNAs. ds32 is a double-stranded RNA with complete complementarity in the two strands. LE21, a single-stranded RNA with minimal intramolecular base pairing, was used to generate ds34. The amount of each input RNA was normalized to 100%. SL13, a 13-nt RNA was produced by in vitro transcription by use of T7 polymerase. 5′-end phosphate was replaced with radiolabeled phosphate by sequential treatment with alkaline phosphatase and T4 polynucleotide kinase. Radiolabeled RNA was gel purified before use.
