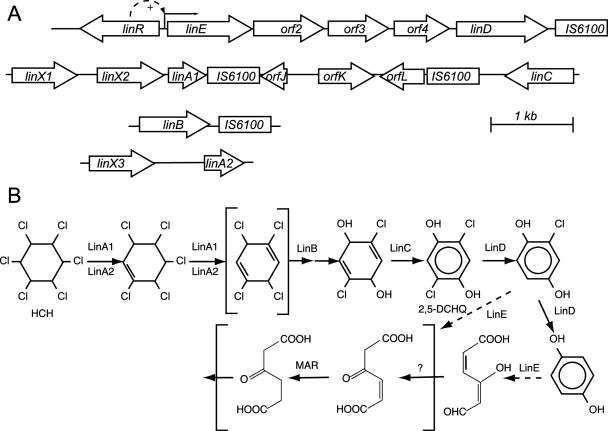
FIG. 1.
Organization of the lin genes in S. paucimobilis strain B90A. (A) Organization of the relevant regions analyzed by DNA sequencing with the lin genes indicated (5). The only known regulatable promoter among the lin genes is in front of linE and is activated by LinR (21). The function of the linX gene product is not known, but based on sequence homology, it was predicted to be a dehydrogenase perhaps similar in function to LinC (24). (B) Simplified version of the degradation pathway for γ-HCH (http://umbbd.ahc.umn.edu/ghch/ghch_image_map.html). Reactions catalyzed by Lin enzymes are indicated as such. When more than one arrow is drawn, reactions are supposed to have intermediate steps involving unstable intermediates. The part shown within brackets remains speculative and is based on analogies to maleylacetate degradation. 2,5-DCHQ, 2,5-dichlorohydroquinone; MAR, maleylacetate reductase.