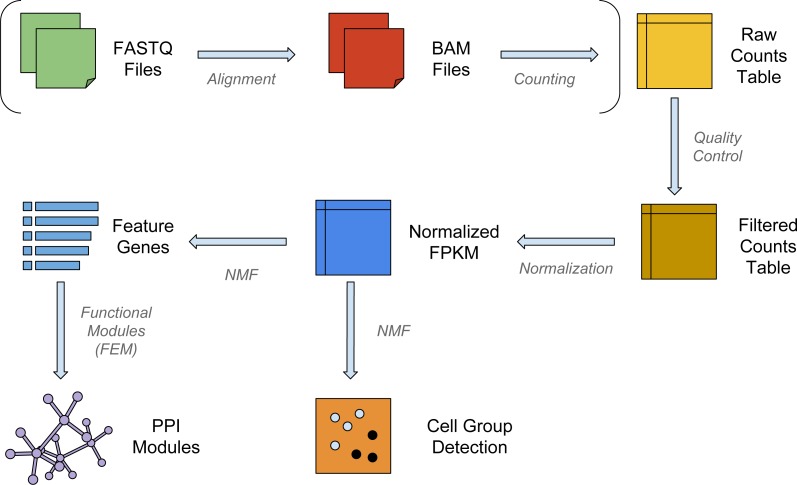
Figure 1. The workflow of NMFEM.
The input can be either FASTQ files or a raw counts table. If FASTQ files are used, they are aligned using TopHat and counted using FeatureCounts (steps shown in brackets). The input or calculated raw counts table are filtered by samples and genes, converted into FPKMs using gene lengths, and normalized by samples. We then run NMF method on them to detect groups of cells, and find the feature genes separating the detected groups. Finally, we feed the feature genes as seed genes in FEM, and generate PPI gene modules that contain highly differentially expressed genes.