Abstract
Paracoccidioides brasiliensis, a thermodimorphic fungus, is the causative agent of the prevalent systemic mycosis in Latin America, paracoccidioidomycosis (PCM). Here, we describe the microsatellite patterns observed in a collection of P. brasiliensis random sequence tags. We identified 1,117 microsatellite patterns in about 3.8 Mb of unique sequences (0.47% of the total DNA used in the analysis). The majority of these microsatellites (87.5%) are found in noncoding sequences. We used two polymorphic microsatellites located on noncoding and coding sequences, as well as two microsatellites located on introns, as molecular markers to discriminate P. brasiliensis isolates, to look for relationships between the genetic background of the strains and the types of human disease they cause. We did not observe any correlation between the clinical form of human PCM and four simple sequence repeat patterns analyzed.
Paracoccidioides brasiliensis, a thermodimorphic fungus, is the causative agent of the prevalent systemic mycosis in Latin America, paracoccidioidomycosis (PCM). Epidemiological data indicate a broad geographic distribution of P. brasiliensis in Central and South America, from Mexico to Argentina (31). The pathogen apparently has its natural habitat in soil or in plants in areas where PCM is endemic, and rural workers appear to become infected by inhaling dust containing the infecting propagules (32). It is estimated that as many as 10 million individuals could be infected with P. brasiliensis, acquired by inhalation of airborne microconidia, which reach the pulmonary alveolar epithelium and transform into the parasitic yeast form (27). The human form of (PCM) caused by this fungus is characterized by a range of clinical manifestations from benign or asymptomatic forms to severe and disseminated disease that is often fatal. The development of PCM depends on interactions between fungus and host components. Many authors have tried to correlate certain characteristics of P. brasiliensis isolates with virulence without success (22, 37, 42, 44). In P. brasiliensis, restriction fragment length polymorphism and random amplified polymorphic DNA (RAPD) markers have been used in attempts to establish epidemiological and phylogenetic relationships between isolates that have different degrees of virulence and that come from distinct geographical regions (3, 23, 25, 26). Although, it has been proposed that P. brasiliensis isolates differ in their ability to cause human disease (36), the issue is far from settled (25). Recently, Hebeler-Barbosa et al. (14) completed the first genetic analysis of P. brasiliensis isolates from 10 armadillos and confirmed their similarity with 19 clinical isolates by DNA sequencing. These authors showed by sequence comparison of the internal transcribed spacer 1 and internal transcribed spacer 2 regions that eight isolates differed by one or three sites among the five polymorphic sites found, suggesting the existence of two genetic groups.
The main antigenic component described in P. brasiliensis is gp43, an exocellular glycoprotein containing a single oligosaccharide chain; it elicits a strong humoral response and can be detected in PCM patient serum (for a review, see reference 41). gp43 is a potential virulence factor because it binds murine laminin, resulting in increased pathogenicity of yeast cells (43). Morais et al. (24) reported P. brasiliensis gp43 gene polymorphism in a variety of P. brasiliensis isolates from patients suffering from chronic and acute PCM. These authors observed that the P. brasiliensis gp43 gene sequences of three isolates from patients with pulmonary or chronic PCM were phylogenetically distant from the sequences of other isolates. These results suggest a possible correlation between P. brasiliensis gp43 gene polymorphism and the degree of pathogenicity of these strains in the animal model.
Microsatellites or simple sequence repeats (SSRs) are tandemly repeated tracts of DNA composed of 1- to 6-bp-long units. They are omnipresent in prokaryotes and eukaryotes, even in the smallest bacterial genomes, and are found anywhere in the genome in both protein-encoding and noncoding regions (40). SSRs are considered to be evolutionarily neutral DNA markers (20). Length polymorphism arises from variations in the number of repeated units, probably due to DNA polymerase slippage during the replication of SSRs (19). They have been used for both population genetics and typing studies because they have several advantages as markers, such as that they are highly polymorphic, multiallelic, highly reproducible, and detectable by PCR (29).
Recently, we established a collection of about 3.8 Mb of unique random sequence tags (RSTs) (M. P. Nobrega et al., unpublished data) and decided to investigate the microsatellite occurrence in this set. Here, we describe the microsatellite patterns observed and use some of them as molecular markers to discriminate P. brasiliensis isolates in a search for correlations between the genetic background of the strains and the types of human disease they cause.
MATERIALS AND METHODS
Fungal strains and DNA preparation.
We analyzed 23 isolates (4 environmental isolates, 18 clinical isolates, and the Pb18 isolate) (Pb18 was kindly provided by Z. P. Camargo, Universidade Federal de Saõ Paulo, Brazil) that are listed in Table 1. Yeast cells were grown to the logarithmic phase in 125-ml Erlenmeyer flasks containing 25 ml of Fava-Neto's medium as previously described (35) at 37°C with constant shaking. DNA was prepared according to a glass bead protocol (33).
TABLE 1.
P. brasiliensis isolates analyzed in this study from patients located in São Paulo and south of the Minas Gerais State, Brazil
| Clinical isolate | Isolation date (mo/day/yr) | Classification | Material |
|---|---|---|---|
| Pb18 | |||
| Pb51 | 8/27/99 | Chronic | Skin biopsy |
| Pb52 | 8/30/99 | Acute/HIV | Skin biopsy |
| Pb61 | 9/24/00 | Acute/HIV | Skin biopsy |
| Pb66 | 3/15/01 | Chronic | Skin biopsy |
| Pb67 | 4/18/01 | Acute/HIV | Mouth lesion |
| Pb71 | 8/08/01 | Acute | Abscess |
| Pb72 | 7/04/01 | Chronic | Skin biopsy |
| Pb74 | 7/30/01 | Chronic | Bronchoalveolar lavage |
| Pb76 | 1/11/02 | Acute/HIV | Duodenal biopsy |
| Pb78 | 1/15/02 | Acute/HIV | Sputum |
| Pb79 | 2/12/02 | Chronic | Sputum |
| Pb80 | 9/27/01 | Acute | Skin biopsy |
| Pb82 | 11/29/01 | Chronic | Lymph node |
| Pb85 | 9/25/01 | Acute | Abscess |
| Pb89 | 3/28/02 | Acute | Blood |
| Pb91 | 9/24/02 | Chronic | Sputum |
| Pb92 | 11/21/02 | Acute | Blood |
| Pb93 | 12/02/02 | Chronic | Peritoneal fluid |
| PbIbiá | 6/97 | Soil from Ibiá region, Minas Gerais State | |
| PbT1 | 6/99 | Armadillo from Ibiá region, Minas Gerais State | |
| PbT2 | 6/99 | Armadillo from Ibiá region, Minas Gerais State | |
| PbT3 | 6/99 | Armadillo from Ibiá region, Minas Gerais State |
PCRs.
Using Primer Express design software, version 1.0 (Applied Biosystems), we designed PCR primers for amplifying each DNA fragment that contains microsatellites. The 40-μl amplification mixture included 1× Taq DNA platinum buffer (Invitrogen), 0.5 μM of each primer (Table 2), a 0.2 mM deoxynucleotide triphosphate mixture, 2.5 U of Taq DNA platinum polymerase (Invitrogen), and 100 ng of genomic DNA. PCR amplification was carried out with a PTC100 96-well thermal cycler (MJ Research) at 95°C for 1 min; for 38 cycles at 95°C for 1 min, 39 to 54.4°C (depending on the fragment) for 1 min, and 72°C for 1 min; and followed by an extension step at 72°C for 10 min. After the reaction, the PCR products were purified with a QIAGEN PCR cleanup kit, following the manufacturer's instructions. Sequencing reactions were prepared with the BigDye Terminator Cycle Sequencing kit (Applied Biosystems), with the primers listed in Table 2. The nucleotide sequences in both strands were determined by primer elongation with an ABI3100 automated DNA sequencer (Applied Biosystems).
TABLE 2.
List of primers used in this work
| Primer | Sequence | Amplified SSR (bp) | NCBI accession no. |
|---|---|---|---|
| MS5 | 5′-TGCCCGAAGCAGCCCCCCGGG-3′ | ATTT (236) | BQ503230 |
| MS6 | 5′-GAGAAAGTGAGTTGGTTTACG-3′ | ||
| MS11 | 5′-TTTGCTACACTTCCCTCTCCC-3′ | AT (410) | CL524685 |
| MS12 | 5′-CTTCCCCCATTCTGATTCTCG-3′ | ||
| MG13 | 5′-CACGTGTCAAGTCATAATAAATAG-3′ | AT, ATTT (251) | CL524686 |
| MG14 | 5′-AATCTGCTGCCAATAGTCAT-3′ | ||
| MG15 | 5′-GCACAGACGCAAAATATGC-3′ | TAAA (152) | CL524687 |
| MG16 | 5′-GGTGGAAAAAGATATGCGAA-3′ | ||
| MG21 | 5′-AACAATCAAACCGGGAG-3′ | AAAAGG (214) | CL524688 |
| MG22 | 5′-GGAGGGATAGGAACGAATT-3′ | ||
| MGI125 | 5′-TCAAACTGACAACCTCAGCC-3′ | TCA (203) | CL524689 |
| MGI126 | 5′-TAAGAAGATGGATGAGGCCC-3′ | ||
| MGI127 | 5′-AAAAATAACTACGCAGACGC-3′ | CCCA (251) | CL524690 |
| MGI128 | 5′-TTAAGCTGGGCTTTGGGTAC-3′ |
Data handling and analysis.
A pipeline was built to analyze and assemble the P. brasiliensis RST sequences. Sequences were automatically edited for each RST with the programs Phred-Phrap (4, 5), Consed (12), and Crossmatch from Phrap (13). The sequences were cleaned from the pUC18 vector sequences with Crossmatch (13); RSTs with a quality value of at least 20 were considered for further analysis. Edited sequences were clustered with the Phrap program (5). To identify if the microsatellite was at either a coding or noncoding region, the P. brasiliensis clusters containing microsatellites were compared with the BLASTX and BLASTN algorithms (1) with the the National Center for Biotechnology Information (NCBI) nonredundant database (http://ncbi.nlm.nih.gov/), several fungal genome databases (www.broad.mit.edu), and the P. brasiliensis expressed sequence tag project databases (http://143.107.203.68/est/default.html). When E values greater than 10−5 were obtained, they were considered not statistically significant (no significant match).
We have extracted all the microsatellites from our RST databank (Table 3) by using the methodology described by Jurka and Pethiyagoda (16). Essentially, we extracted only simple repeats composed of tandemly repeated basic units 1 to 6 nucleotides (nt) long. Most simple repeats and their complementary counterparts can be represented by several different basic unit patterns. For example, the pattern (GCC)n listed by its unit name GCC in Table 3, also represents (CCG)n, (CGC)n, (GGC)n, (GCG)n, and (CGG)n. Each simple sequence was counted on one strand only and consequently the length is given by the number of nucleotides. Furthermore, whenever tandemly repeated patterns with different unit sizes were identical, they were listed under the smallest unit size. For example, patterns like (ACACAC)n or (ACAC)n were included into the category (AC)n. As a result, the total number of theoretically possible, nonoverlapping patterns was reduced to 501 (2 monomeric, 4 dimeric, 10 trimeric, 33 tetrameric, 102 pentameric, and 350 hexameric patterns).
TABLE 3.
Summary of the data for 67 microsatellite patterns with two occurrences and with two or more sizes
| Msata | Maximumb | Meanc | SD | Frequencyd | Repeatse | Total lengthf | Abundance (%)g |
|---|---|---|---|---|---|---|---|
| A | 39 | 17.1 | 6.0 | 149 | 23 | 2,552 | 14.1 |
| C | 20 | 14.2 | 1.9 | 34 | 7 | 483 | 2.6 |
| AC | 74 | 25.0 | 14.1 | 28 | 14 | 702 | 3.8 |
| AG | 68 | 20.2 | 11.2 | 32 | 12 | 648 | 3.5 |
| AT | 40 | 17.8 | 6.1 | 110 | 12 | 1,958 | 10.8 |
| ACG | 24 | 17 | 4.4 | 6 | 5 | 102 | 0.5 |
| CAA | 39 | 17.3 | 6.6 | 23 | 6 | 399 | 2.2 |
| CCA | 21 | 14.8 | 2.4 | 17 | 4 | 252 | 1.3 |
| CTA | 36 | 24 | 12 | 2 | 2 | 48 | 0.2 |
| GAA | 54 | 20 | 12.3 | 23 | 8 | 462 | 2.5 |
| GCA | 18 | 13.3 | 2.2 | 24 | 3 | 321 | 1.7 |
| GCC | 15 | 13.5 | 1.5 | 4 | 2 | 54 | 0.3 |
| GGA | 18 | 13.6 | 1.9 | 11 | 3 | 150 | 0.8 |
| TAA | 108 | 31.2 | 26.5 | 30 | 14 | 936 | 5.1 |
| TCA | 51 | 18.8 | 9.0 | 22 | 6 | 414 | 2.2 |
| AACC | 32 | 14.4 | 5.9 | 10 | 3 | 144 | 0.8 |
| AACT | 20 | 12.7 | 1.9 | 21 | 3 | 268 | 1.4 |
| ATGC | 20 | 13.3 | 2.9 | 6 | 2 | 80 | 0.4 |
| CAAA | 24 | 21.3 | 3.7 | 3 | 2 | 64 | 0.3 |
| GAAA | 44 | 14.3 | 6.4 | 24 | 3 | 344 | 1.9 |
| GACA | 40 | 24 | 11.7 | 3 | 3 | 72 | 0.4 |
| GCCA | 24 | 15 | 5.1 | 4 | 2 | 60 | 0.3 |
| GGGA | 20 | 14 | 3.4 | 4 | 2 | 56 | 0.3 |
| TAAA | 36 | 16.3 | 6.7 | 24 | 6 | 392 | 2.1 |
| TACA | 20 | 13.6 | 2.6 | 10 | 3 | 136 | 0.7 |
| TCAA | 16 | 12.8 | 1.6 | 5 | 2 | 64 | 0.3 |
| TCCA | 20 | 15 | 3.3 | 4 | 3 | 60 | 0.3 |
| TGAA | 20 | 14.6 | 3.7 | 3 | 2 | 44 | 0.2 |
| ACAGC | 20 | 17.5 | 2.5 | 2 | 2 | 35 | 0.1 |
| ATGCC | 30 | 22.5 | 7.5 | 2 | 2 | 45 | 0.2 |
| CAAAA | 30 | 22.5 | 7.5 | 2 | 2 | 45 | 0.2 |
| GAAAA | 40 | 19.8 | 6.4 | 25 | 5 | 495 | 2.7 |
| TAAAA | 35 | 18 | 5.0 | 15 | 3 | 270 | 1.4 |
| TTAAA | 20 | 17.5 | 2.5 | 2 | 2 | 35 | 0.1 |
| AAAAGG | 42 | 15.3 | 9.4 | 9 | 2 | 138 | 0.7 |
| AAAATC | 24 | 15 | 5.1 | 4 | 2 | 60 | 0.3 |
| AAAATT | 18 | 14 | 2.8 | 3 | 2 | 42 | 0.2 |
| AAAGAG | 30 | 14.5 | 6.2 | 7 | 2 | 102 | 0.5 |
| AAAGGG | 18 | 15 | 3 | 2 | 2 | 30 | 0.1 |
| AAATAG | 18 | 15 | 3 | 2 | 2 | 30 | 0.1 |
| AAATAT | 18 | 13.5 | 2.5 | 4 | 2 | 54 | 0.3 |
| AAATGG | 18 | 13 | 2.2 | 6 | 2 | 78 | 0.4 |
| AACAGC | 18 | 13.7 | 2.7 | 7 | 2 | 96 | 0.5 |
| AACCAC | 30 | 19.5 | 7.7 | 4 | 3 | 78 | 0.4 |
| AACCGC | 24 | 15 | 5.1 | 4 | 2 | 60 | 0.3 |
| AACCTC | 30 | 24 | 6 | 2 | 2 | 48 | 0.2 |
| AACGAC | 18 | 14 | 2.8 | 3 | 2 | 42 | 0.2 |
| AAGATG | 18 | 14 | 2.8 | 3 | 2 | 42 | 0.2 |
| AATAAC | 18 | 13.5 | 2.5 | 4 | 2 | 54 | 0.3 |
| AATATT | 18 | 14 | 2.8 | 3 | 2 | 42 | 0.2 |
| AATCTC | 18 | 13.2 | 2.4 | 5 | 2 | 66 | 0.3 |
| AATTAT | 24 | 16 | 5.6 | 3 | 2 | 48 | 0.2 |
| ACACCC | 18 | 14 | 2.8 | 3 | 2 | 42 | 0.2 |
| ACACTC | 24 | 18 | 6 | 2 | 2 | 36 | 0.2 |
| ACCGGG | 18 | 15 | 3 | 2 | 2 | 30 | 0.1 |
| ACGAGG | 18 | 15 | 3 | 2 | 2 | 30 | 0.1 |
| ACTGCT | 30 | 20 | 7.4 | 3 | 3 | 60 | 0.3 |
| ATATCC | 24 | 20 | 2.8 | 3 | 2 | 60 | 0.3 |
| ATATGC | 18 | 15 | 3 | 2 | 2 | 30 | 0.1 |
| ATCCAC | 18 | 12.8 | 2.0 | 7 | 2 | 90 | 0.5 |
| ATCCGC | 24 | 15 | 5.1 | 4 | 2 | 60 | 0.3 |
| ATCGCC | 18 | 15 | 3 | 2 | 2 | 30 | 0.1 |
| ATCTCC | 18 | 14 | 2.8 | 3 | 2 | 42 | 0.2 |
| CCCCAA | 36 | 20.4 | 8.9 | 5 | 4 | 102 | 0.5 |
| GAAAAA | 24 | 12.6 | 2.4 | 27 | 3 | 342 | 1.8 |
| TAAAAA | 30 | 14.8 | 5.3 | 25 | 4 | 372 | 2.0 |
| TATACA | 18 | 14.4 | 2.9 | 5 | 2 | 72 | 0.4 |
Of a total of 501 patterns, 58 have two occurrences and a single size, 101 have a single occurrence, and 275 patterns were not detected. Msat, type of basic pattern in the simple repeat.
Maximum length of segments.
Average length of segments.
The number of times the pattern appears independently of the length of the size of the segment.
Number of different length ranges (Ne) found among repeats longer than 12 nt. For example, CAAA and its complement are represented by two fragments (Ne = 2): one is 12 nt long and the other is 24 nt long. (AC)n is present in 14 different length fragments (Ne = 14). Only patterns with N ≥ 2 are shown.
Total length of repeat units over 12 nt long.
Relative abundance of repeats over 12 nt long.
Phylogenetic analysis was carried out with the MEGA2 (Molecular Evolutionary Genetics Analysis) software, version 2.1 (http://www.megasoftware.net; 18). The SSR sequences were aligned and the dendrogram was determined by using the ClustalX and the neighbor-joining method, respectively (30, 39). A bootstrap analysis (6) was performed (for 1,025 repeats) to evaluate the topology of the phylogenetic tree.
RESULTS
Identification of microsatellites.
We used the methodology described by Jurka and Pethiyagoda (16) to identify microsatellite patterns in a collection of 6,689 clusters of RSTs that correspond to 3.8 Mb (Nobrega et al., unpublished; http://143.107.203.68/rstpb/frame2.htm). We selected repeat segments over 12 nt long and represented by more than a length size in the P. brasiliensis Pb18 isolate RST database. The SSR patterns range from 1 to 6 nt. The SSRs represent about 0.47% of the total DNA in the analysis. Of 501 possible types of simple repeats, only 125 met these criteria. Table 3 shows 67 of these microsatellite patterns that displayed two occurrences and two or more different sizes.
According to Jurka and Pethiyagoda (16), a possible indicator of sequence variability in microsatellites is the maximal observed lengths per given microsatellite pattern (Table 3, columns “Frequency” and “Repeats”). These numbers must also be used separately for each group of patterns (monomeric, dimeric, etc.) to reduce the overall impact of sample sizes on their absolute values. In Table 3, the column “Abundance” is equivalent to the column “Total length,” as the former lists the proportions of each individual pattern that are >12 nt long relative to the sum of all pattern lengths of ≥12 nt listed in the column “Total length.” Overall, the most abundant are hexanucleotide repeats representing 29.4% of the total simple repeats, followed by dinucleotide (18.3%), trinucleotide (17.3%), mononucleotide (16.8%), tetranucleotide (11.8%), and pentanucleotide (6.4%) repeats. Noticeably, pentanucleotide repeats are underrepresented in our database when compared to the others. As can be seen from Table 3 (columns “Frequency” and “Repeats”), TAA repeats are probably more polymorphic than GCC repeats because they have 108 nt, as opposed to 15 nt (the longest observable length), and 14, as opposed to 2, types of length size. The same can be observed with TAAA when compared to TCAA, where 36 nt as opposed to 16 nt is the longest observable length, and there are six, as opposed to two, types of length size.
Comparison of the genomic sequence that comprises a specific microsatellite plus 200 bp upstream and downstream from the microsatellite, against a collection of 4,692 P. brasiliensis expressed sequence tags (11), the NCBI databank (http://ncbi.nlm.nih.gov/), and fungal genome databases (www.broad.mit.edu) allowed us to summarize the genomic distribution of the P. brasiliensis microsatellites (Table 4). There are 887 clusters with microsatellites and 1,117 clusters with more than one SSR, because some sequences carry more than one microsatellite pattern (Table 4). Most of these microsatellite patterns (87.5%) are found in noncoding sequences, 10.9% are found in coding sequences, 1.3% are found in intron sequences, and 0.3% are located in transposons (Table 4).
TABLE 4.
Main features of seven clusters chosen for microsatellite characterization
| Microsatellite primer paira | Microsatellite pattern (bp)b | No. of repeats at microsatellite locusc | Microsatellite featuresd |
|---|---|---|---|
| MS5XMS6 | TAAA (236) | 4-11 | Coding region; NP_587920.1, probable 6-phosphogluconolactonase Schizosacharomyces pombe; 3e-64/52/66 |
| MS11XMS12 | AT (410) | 10-14 | Noncoding region |
| MG13XMG14 | AT, ATTT, AT (251) | 9-19, 1-6, 2-20 | Located in a Noncoding region |
| MG15XMG16 | TAAA (152) | 3-6 | Noncoding region |
| MG21XMG22 | AAAAGG (214) | 1-8 | coding region; EAA60777 hypothetical protein AN4735.2, Aspergillus nidulans; 5e-14/70/88 |
| MGI25XMGI26 | TCA (203) | 4-5 | Intron region; EAA61572.1 hypothetical protein AN7784.2, Aspergillus nidulans; 5e-19/50/62 |
| MGI27XMGI28 | CCCA (251) | 3 | Intron region; EAA28067.1 hypothetical protein, Neurospora crassa; 5e-36/61/66 |
Each primer pair is shown with an X linking the two primers (e.g., MS5 and MS6).
Size of DNA fragment amplified by PCR in strain Pb18.
Range in the number of simple repeats in the clinical and environmental isolates listed in Table 1.
Features are listed as follows: microsatellite location; NCBI accession number, homologue; E-value/% identity/% similarity.
Characterization of microsatellite loci.
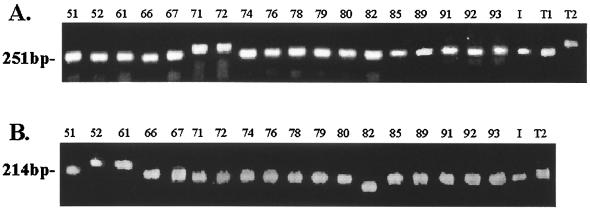
Based mainly on the criterium of sequence variability proposed above, we chose seven clusters that contained microsatellites to analyze their degrees of polymorphism. Two of the corresponding microsatellite patterns were located on coding sequences (TAAA and AAAAGG), three were on noncoding sequences (AT, AT/ATTT/AT, and TAAA), and two were in intron sequences (TCA and CCCA) (Table 4). We designed oligonucleotide primers (Table 2) based on the sequences that limit these microsatellite patterns and PCR amplified the DNA fragments. Size distribution and sequence analysis of several clinical and environmental isolates (Table 1) revealed that clusters MG13XMG14 (consisting of MG13 and MG14) and MG21XMG22 have the most polymorphic microsatellite patterns (Fig. 1 and Table 5). The MG13XMG14 cluster has three different microsatellite patterns (AT, ATTT, and another AT) that can range from 6 to 19, 1 to 6, and 2 to 20 repeats, respectively (Table 5). The MG21XMG22 cluster has an AAAAGG simple repeat that can range from 1 to 8 repeats (Tables 4 and 5).
FIG. 1.
Agarose gel (2.0%) showing the PCR amplification products of the corresponding DNA fragments from microsatellite primer pair MG13XMG14.The molecular weights indicated at the left refer to the PCR product amplified from strain Pb18 (A) and primer pair MG21XMG22 (B). The numbers and letters refer to the different isolates described in Table 1. I, PbIbiá; T1, PbT1; and T2, PbT2.
TABLE 5.
Repeats in microsatellites MG13XMG14 and MG21XMG22 of P. brasiliensis
| Isolate | No. of repeats in:
|
|||
|---|---|---|---|---|
| MG13XMG14
|
MG21XMG22a
|
|||
| AT | ATTT | AT | AAAAGG | |
| Pb18 | 10 | 6 | 14 | 7 |
| Pb51 | 15 | 2 | 16 | 2 |
| Pb52 | 14 | 4 | 16 | 7 |
| Pb61 | 18 | 3 | 20 | 8 |
| Pb66 | 10 | 6 | 14 | ND |
| Pb67 | 18 | 3 | 16 | 7 |
| Pb71 | 15 | 4 | 16 | 7 |
| Pb72 | 15 | 4 | 13 | 7 |
| Pb74 | 11 | 6 | 14 | 7 |
| Pb76 | 13 | 4 | 16 | 7 |
| Pb78 | 16 | 4 | 16 | 7 |
| Pb79 | 17 | 3 | 16 | ND |
| Pb80 | 11 | 1 | 2 | 5 |
| Pb82 | 16 | 4 | 17 | ND |
| Pb85 | 17 | 3 | 16 | 1 |
| Pb89 | 15 | 4 | 16 | ND |
| Pb91 | 19 | 2 | 17 | 3 |
| Pb92 | 18 | 3 | 16 | 7 |
| Pb93 | 19 | 3 | 17 | 2 |
| PbIbiá | 17 | 4 | 16 | 3 |
| PbT1 | 15 | 4 | 14 | 6 |
| PbT2 | 19 | 1 | 2 | 7 |
| PbT3 | 9 | 1 | 2 | 7 |
ND, not determined.
Use of microsatellites as markers to evaluate correlation between genetic background of strains and virulence degree.
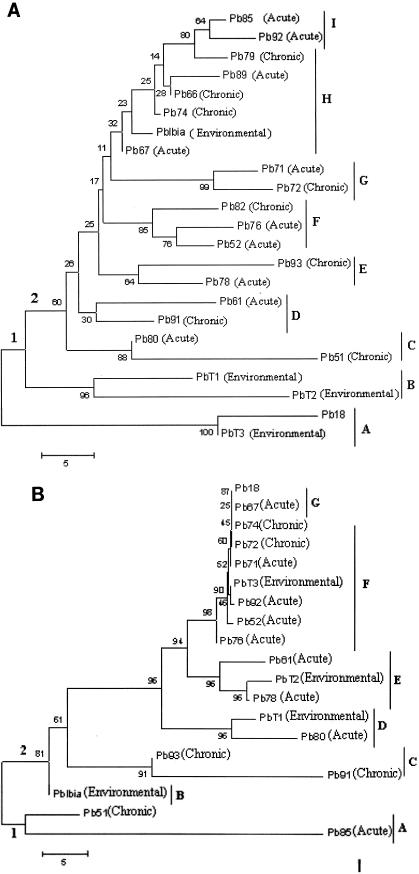
We have used these two most polymorphic clusters (MG13XMG14 and MG21XMG22) as well as the DNA sequences of the two clusters that have microsatellite patterns in the introns (MGI25XMGI26 and MGI27XMGI28) to evaluate relatedness among clinical and environmental isolates of P. brasiliensis. The clinical isolates were different from each other in the type of human disease they caused: 8 caused chronic disease, and 10 caused acute disease (Table 1). In addition, four isolates were environmental isolates: three were isolated from armadillos, and one was isolated from soil (Table 1). Two phylogenetic trees were constructed by the neighbor-joining method based on the number of repeats obtained by PCR amplification of the microsatellites MG13XMG14 and MG21XMG22 (Fig. 2). All isolates had different multilocus genotypes, but there was no clustering of isolates associated with chronic or acute disease or with the environment. The number of repeats was randomly distributed among clinical versus nonclinical isolates (Table 5). The reliability of the phylogenetic trees inferred was verified by the bootstrap method (Fig. 2). In the first tree (MG13XMG14) (Fig. 2A), two main groups were identified. (i) In the first group, three environmental isolates (PbIbiá, PbT1, and PbT3) clustered in two different subgroups (A and B). (ii) In the second group, there are seven subgroups (C to I) which have not shown clustering between isolates that cause chronic or acute disease. Furthermore, the fourth environmental isolate in the second group clustered with isolates that cause chronic or acute disease (subgroup H). In the second tree (MG21XMG22) (Fig. 2B), two main groups were also observed. (i) In the first group, one environmental isolate, an isolate causing chronic disease, and an isolate causing acute disease (PbIbiá, Pb51, and Pb85) clustered into two different subgroups (A and B). (ii) In the second group, there are seven subgroups (C to G) which have not shown clustering between isolates that cause chronic or acute disease. Comparable results were observed for the microsatellite clusters MGI25XMGI26 and MGI27XMGI28 (data not shown). Taken together, these results suggest there is no clear correlation between the genetic background of the isolates, as measured here, and the types of human disease they cause.
FIG.2.
Phylogenetic trees based on the MG13XMG14 (A) and MG21XMG22 (B) microsatellite sequences show the relationships of the 23 P. brasiliensis isolates. These unrooted trees were constructed by the neighbor-joining method. Topology was also evaluated by bootstrap analysis (MEGA2 program; 1,025 repeats). The numerical values in the trees represent bootstrap results. The distance between two strains is the sum of the branch lengths between them. The identified subgroups are indicated by the letters at the right.
DISCUSSION
The main objective of this study was to develop microsatellite or SSR markers for the pathogenic dimorphic fungus P. brasiliensis. To identify SSR markers, we digitally screened an RST collection composed of 3.8 Mb of unique sequences, which should represent about 15 to 20% of the P. brasiliensis genome. SSR markers have been successfully applied for typing, mapping, and population studies of several fungal species, such as Saccharomyces cerevisiae, Candida spp., Aspergillus fumigatus, Magnaporthe grisea, Coccidioides spp., and Histoplasma capsulatum (2, 7-10, 15, 17, 21, 28, 34, 38). SSRs constitute a rather large fraction of noncoding DNA and are relatively rare in protein-encoding regions (20). Accordingly, most of the observed P. brasiliensis SSRs are located in such noncoding regions. The majority of SSRs found in many species are dinucleotides; in primates, mononucleotides [mainly poly(A-T) tracts] are the most copious classes of SSRs (20). We have observed that the most abundant SSRs in P. brasiliensis are hexanucleotides (about 29%). More fungal genomes are being sequenced (see the White Paper Initiative at www.wi.mit.edu), which will provide more information about the type, length, and frequency of SSRs in fungi.
P. brasiliensis usually reaches the human host through the respiratory route by inhalation of airborne mycelial propagules that convert to the tissue yeast form, initiating infection; with time, infection may give rise to clinical PCM, a disease that may adopt different clinical forms (31). Once established, the disease may be acute or chronic, depending on the severity and localization of lesions. Variations in the intensity, extension, dissemination, and characteristics of the lesions in PCM will occur in a given patient depending on fungal virulence, fluctuations of the host defense mechanisms, and environmental factors (23, 32). In the present work, we evaluate SSR profiles as genetically associated elements with the potential to discriminate P. brasiliensis isolates according to their degree of virulence as determined from the corresponding clinical forms of patients with PCM. In typing with SSRs, an entire stretch of sequence is surveyed for genetic variation through length polymorphisms. Recently, Calcagno et al. (3) using RAPD analysis demonstrated that genetic differentiation could be associated with geographical region but not with different clinical manifestations of human PCM. Motta et al. (25) found no correlation between RAPD patterns and the type of pathology as observed with experimental infection in mice or in the clinical form of human PCM. In contrast with these authors, Molinari-Madlum et al. (23) have shown that RAPD patterns correlated with the virulence degree of P. brasiliensis isolates. Nevertheless, our results are similar to those of Motta et al. (25) and Calcagno et al. (3); we observed no correlation between the clinical form of human PCM and four SSR patterns. Interestingly, the P. brasiliensis isolates derived from AIDS patients (in this work, these isolates are classified as isolates that cause acute disease) have also not clustered, suggesting the possibility that any P. brasiliensis isolate could behave as an opportunistic pathogen in immunocompromised HIV patients.
The P. brasiliensis SSRs now available will provide new opportunities for epidemiogical and phylogenetic studies of this organism.
Acknowledgments
We thank the following agencies for their financial support: Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), both of Brazil (to M.H.S.G., M.P.N., F.G.N., and G.H.G.), and NIH Fogarty International Center grant R03TW001308 (to J.W.T. and J.G.M.).
We also thank Leila M. Toffoli for expert technical help and Diógenes Custódio Duarte Ribeiro for help with the bioinformatics.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bart-Delabesse, E., J. F. Humbert, E. Delabesse, and S. Bretagne. 1998. Microsatellite markers for typing Aspergillus fumigatus isolates. J. Clin. Microbiol. 36:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calcagno, A. M., G. Niño-Vega, F. San-Blas, and G. San-Blas. 1998. Geographic discrimination of Paracoccidioides brasiliensis strains by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 36:1733-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 5.Ewing, B., and P. Green. 1998. Base calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 7.Fisher, M. C., G. Koenig, T. J. White, and J. W. Taylor. 2000. A test for concordance between the multilocus genealogies of genes and microsatellites in the pathogenic fungus Coccidioides immitis. Mol. Biol. Evol. 17:1164-1174. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, M. C., G. L. Koenig, T. J. White, G. San-Blas, R. Negroni, I. G. Alvarez, B. Wanke, and J. W. Taylor. 2001. Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc. Natl. Acad. Sci. USA 98:4558-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, M. C., G. L. Koening, T. J. White, and J. W. Taylor. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73-84. [PubMed] [Google Scholar]
- 10.Fisher, M. C., B. Rannala, V. Chaturverdi, and J. W. Taylor. 2002. Disease surveillance in recombining pathogens: multilocus genotypes identify sources of Coccidioides infections. Proc. Natl. Acad. USA 99:9067-9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldman, G. H., E. d. R. Marques, D. C. Ribeiro, L. A. de Souza Bernardes, A. C. Quiapin, P. M. Vitorelli, M. Savoldi, C. P. Semighini, R. C. de Oliveira, L. R. Nunes, L. R. Travassos, R. Puccia, W. L. Batista, L. E. Ferreira, J. C. Moreira, A. P. Bogossian, F. Tekaia, M. P. Nobrega, F. G. Nobrega, and M. H. S. Goldman. 2003. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity. Eukaryot. Cell 2:34-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 13.Green, P. 1996. Phred, Phrap, Consed. http://bozeman.mbt.washington.edu/phredphrapconsed.html.
- 14.Hebeler-Barbosa, F., F. V. Morais, M. R. Montenegro, E. E. Kuramae, B. Montes, J. G. McEwen, E. Bagagli, and R. Puccia. 2003. Comparison of the sequences of the internal transcribed spacer regions and PbGP43 genes of Paracoccidioides brasiliensis from patients and armadillos (Dasypus novemcinctus). J. Clin. Microbiol. 41:5735-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennequin, C., A. Thierry, G. F. Richard, G. Lecointre, H. V. Nguyen, C. Gaillardin, and B. Dujon. 2001. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurka, J., and C. Pethiyagoda. 1995. Simple repetitive DNA sequences from primates: compilation and analysis. J. Mol. Evol. 40:120-126. [DOI] [PubMed] [Google Scholar]
- 17.Kaye, C., J. Milazzo, S. Rozenfeld, M.-H. Lebrun, and D. Tarreau. 2003. The development of simple sequence repeat markers for Magnaporthe grisea and their integration into an established genetic linkage map. Fungal Genet. Biol. 40:207-214. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203-221. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y.-C., A. B. Korol, T. Fahima, A. Beiles, and E. Nevo. 2002. Microsatellites: genomic distribution, putative functions and mutational mechanism: a review. Mol. Ecol. 11:2453-2465. [DOI] [PubMed] [Google Scholar]
- 21.Lunel, F. V., L. Licciardello, S. Stefani, H. A. Verbrugh, W. J. Melchers, J. F. Meis, S. Scherer, and A. van Belkum. 1998. Lack of consistent short sequence repeat polymorphisms in genetically homologous colonizing and invasive Candida albicans strains. J. Bacteriol. 180:3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manocha, M. S., G. San-Blas, and S. Centeno. 1980. Lipid composition of Paracoccidioides brasiliensis: possible correlation with virulence of different strains. J. Gen. Microbiol. 117:147-154. [DOI] [PubMed] [Google Scholar]
- 23.Molinari-Madlum, E. E. W. I., M. S. S. Felipe, and C. M. A. Soares. 2000. Virulence of Paracoccidioides brasiliensis isolates can be correlated to groups by random amplified polymorphic DNA analysis. Med. Mycol. 37:269-276. [PubMed] [Google Scholar]
- 24.Morais, F. V., T. F. Barros, M. K. Fukada, P. S. Cisalpino, and R. Puccia. 2000. Polymorphism in the gene coding for the imunodominant antigen gp43 from the pathogenic fungus Paracoccicioides brasiliensis. J. Clin. Microbiol. 38:3960-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motta, T. R., C. A. Moreira-Filho, R. P. Mendes, L. R. Souza, M. F. Sugizak, S. Baueb, V. L. G. Calich, and C. A. C. Vaz. 2002. Evaluation of DNA polymorphisms amplified by arbitrary primers (RAPD) as genetically associated elements to differentiate virulent and non-virulent Paracoccidioides brasiliensis isolates. FEMS Immunol. Med. Microbiol. 33:151-157. [DOI] [PubMed] [Google Scholar]
- 26.Niño-Vega, G. A., A. M. Calcagno, G. San-Blas, F. San-Blas, G. W. Gooday, and N. A. R. Gow. 2000. RFLP analysis reveals marked geographical isolation between strains of Paracoccidioides brasiliensis. Med. Mycol. 38:437-441. [DOI] [PubMed] [Google Scholar]
- 27.Restrepo, A., J. G. McEwen, and E. Castañeda. 2001. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med. Mycol. 39:233-241. [DOI] [PubMed] [Google Scholar]
- 28.Rosehart, K., M. H. Richards, and M. J. Bidochka. 2002. Microsatellite analysis of environmental and clinical isolates of the opportunist fungal pathogen Aspergillus fumigatus. J. Med. Microbiol. 51:1128-1134. [DOI] [PubMed] [Google Scholar]
- 29.Saghai Maroof, M. A., R. M. Biyashev, G. Yang, Q. Zhang, and R. W. Allard. 1994. Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations, and population dynamics. Proc. Natl. Acad. Sci. USA 91:5466-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.San-Blas, G., and G. Niño-Vega. 2001. Paracoccidioides brasiliensis: virulence and host response, p. 205-226. In R. L. Cihlar and R. A. Calderone (ed.), Fungal pathogenesis: principles and clinical applications. Marcel Dekker, Inc., New York, N.Y.
- 32.San-Blas, G., G. Niño-Vega, and T. Iturriaga. 2002. Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 40:225-242. [DOI] [PubMed] [Google Scholar]
- 33.Semighini, C. P., Z. P. De Camargo, R. Puccia, M. H. Goldman, and G. H. Goldman. 2002. Molecular identification of Paracoccidioides brasiliensis by 5′ nuclease assay. Diagn. Microbiol. Infect. Dis. 44:383-386. [DOI] [PubMed] [Google Scholar]
- 34.Shemer, R., Z. Weissman, N. Hashman, and D. Kornitzer. 2001. A highly polymorphic degenerate microsatellite for molecular strain typing of Candida krusei. Microbiology 147:2021-2028. [DOI] [PubMed] [Google Scholar]
- 35.Silva, S. P., M. S. S., M. Pereira, M. O. Azevedo, and C. M. A. Soares. 1999. Phase transition and stage-specific protein synthesis in the dimorphic fungus Paracoccidioides brasiliensis. Exp. Mycol. 18:294-299. [Google Scholar]
- 36.Soares, C. M. A., E. E. W. I. Mollinari Madlum, S.P. da Silva, M. Pereira, and M. S. S. Felipe. 1995. Characterization of Paracoccidioides brasiliensis isolates by random amplified polymorphic DNA analysis. J. Clin. Microbiol. 33:505-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svidzinsky, T. I. E., and Z. P. Camargo. 1995. Isoenzyme profile of Paracoccidioides brasiliensis. J. Med. Vet. Mycol. 33:281-285. [PubMed] [Google Scholar]
- 38.Taylor, J. W., D. M. Geiser, A. Burt, and V. Koufonapou. 1999. The evolutionary biology and population genetics underlying fungal strain typing. Clin. Microbiol. Rev. 12:126-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tóth, G., Z. Gáspaári, and J. Jurka. 2000. Microosatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10:967-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travassos, L. R., C. P. Taborda, L. K. Iwai, E. C. Cunha-Neto, and R. Puccia. 2004. The gp43 from Paracoccidioides brasiliensis: a major diagnostic antigen and vaccine candidate, p. 279-296. In J. E. Domer and G. S. Kobayashi (ed.), The Mycota XII: human fungal pathogens. Springer-Verlag, Berlin, Germany.
- 42.Vaz, C. A. C., D. W. Mackenzie, V. M. Hearn, Z. P. Camargo, L. M. Singer-Vermes, E. Burger, and V. L. G. Calich. 1994. Gelatinase activity of exoantigens from virulent and non-virulent isolates of Paracoccidioides brasiliensis. J. Med. Vet. Mycol. 32:65-69. [DOI] [PubMed] [Google Scholar]
- 43.Vicentini, A. P., J. L. Gesztesi, M. Franco, W. Souza, J. Moraes, L. R. Travassos, and J. D. Lopes. 1994. Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect. Immun. 62:4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaccharias, D., A. Ueda, M. Moschardi-Bacchi, M. Franco, and G. San-Blas. 1986. A comparative histopathological, immunological and biochemical study of experimental intravenous paracoccidioidomycosis induced in mice by three Paracoccidioides brasiliensis isolates. J. Med. Vet. Mycol. 24:445-454. [PubMed] [Google Scholar]