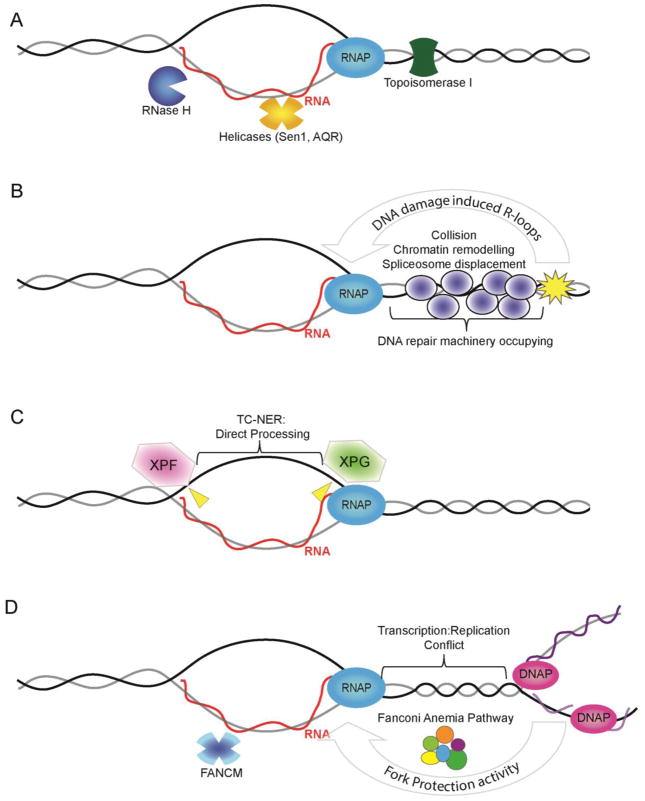
Figure 1.
Potential modes of action for DNA damage proteins in R-loop mitigation. (A) R-loop structure is shown (RNA = red; DNA = grey/black) occurring co-transcriptionally with RNA polymerase (RNAP). The RNA exits RNAPII at a separate position from the template DNA so the RNA must re-invade to form an R-loop. Also shown are core anti-R-loop enzymatic activities: RNase H which degrades the RNA moiety; Helicases such as Senataxin (Sen1)39 or Aquarius (AQR)25 which may unwind R-loops; and Topoisomerase which reduces topological strain favoring RNA invasion. (B) DNA damage induced R-loops. Transcription into a damaged DNA region would experience altered chromatin environment (i.e. remodelers actively suppress transcription next to breaks20), collision with repair proteins or be subject to orchestrated disassembly by DNA repair signaling (e.g. spliceosome displacement favoring R-loops21). (C) Direct processing of R-loops into DNA breaks by the transcription-coupled nucleotide excision repair (TC-NER) machinery. Putative cut sites are shown in yellow although the cut strand is not known25. (D) Interplay between replication fork protection activities and R-loop stability. At sites of transcription-replication conflict the Fanconi Anemia pathway, which includes BRCA1, BRCA2 and XPF, act to suppress R-loop formation36; 37. The FANCM helicase is shown as a mediator that could directly remove R-loops through its strand migration activity36.