Figure 1.
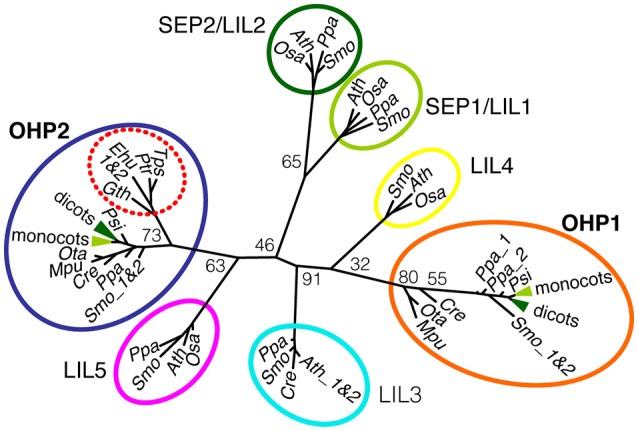
Phylogenetic analysis of OHP1 and OHP2. OHP and SEP/LIL sequences from selected plants and algae were aligned and used to calculate an unrooted maximum likelihood tree (See Supplemental Figure 1 for an excerpt of the alignment). Numbers at branching points are bootstrap values from 100 replicates. OHP1 and OHP2 proteins belong to two evolutionary distinct groups that most likely evolved independently in photosynthetic eukaryotes. Notably, secondary algae in the red lineage only contain OHP2 sequences (red dotted circle) but no nuclear-encoded OHP1. For a better overview, the highly conserved dicot (dark green) and monocot (light green) sequences were collapsed in the OHP1 and OHP2 branches of the tree and SEP/LIL sequences were only included from a small group of green algae and land plants. In addition to the species mentioned, OHP1 and OHP2 sequences from the dicots apple (Malus domestica), barrel clover (Medicago truncatula) and poplar (Populus trichocarpa) were included for the calculation of the tree. Seed plants: Ath, Arabidopsis thaliana; Pop, Populus trichocarpa; Zma, Zea mays, Osa, Oryza sativa; Psi, Picea sitchensis. Lower land plants: Ppa, Physcomitrella patens; Smo, Selaginella moellendorffii. Green algae: Cre, Chlamydonomas reinhardtii; Ota, Ostreococcus tauri; Mpu, Micromonas pusilla. Diatoms: Ptr, Phaeodactylum tricornutum; Tps, Thalassiosira pseudonana. Haptophyte: Ehu, Emiliana huxleyi. Cryptophyte: Gth, Guillardia theta.
