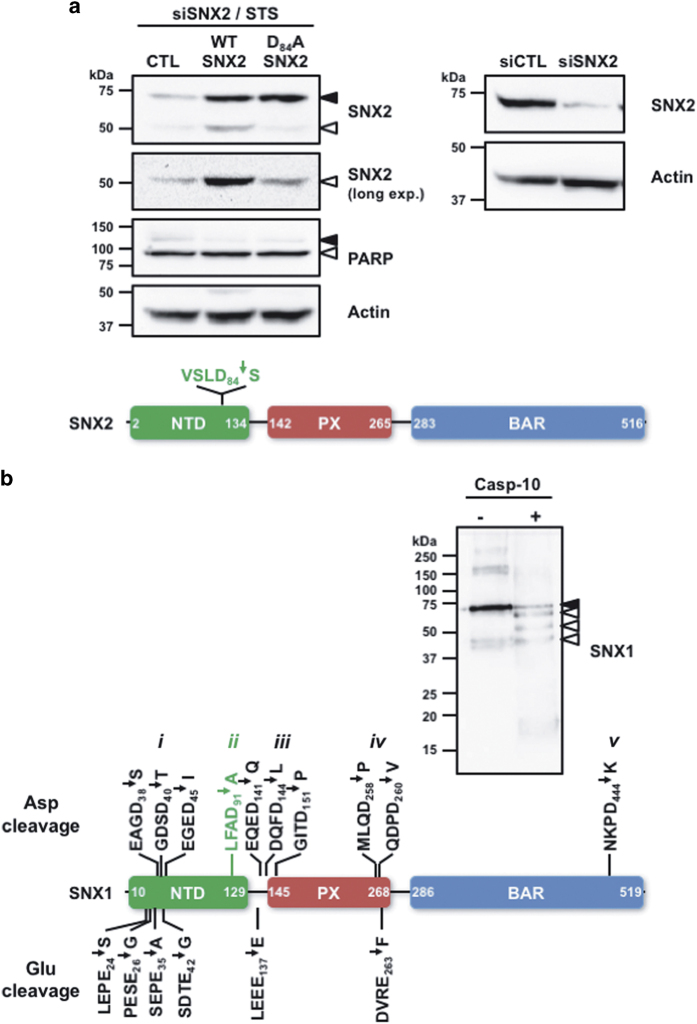
Figure 3.
Identification of caspase cleavage sites in SNX1 and SNX2. (a) HeLa cells were transfected two times at 24 h interval with a SNX2 small interfering RNA (siRNA) and then with either empty (CTL), siRNA-resistant WT or Asp84/Ala SNX2 plasmids. Transfected cells were then incubated with STS (0.5 μM) for 6 h (left panel). These conditions lead to 90.3% SNX2 depletion (right panel). The schematic presents the location of the sole cleavage site found in SNX2. (b) Recombinant SNX1 was incubated with or without recombinant caspase-10 for an hour at 37 °C. An aliquot of each sample was analyzed by immunoblotting using a SNX1 antibody. Samples were analyzed by tandem mass spectrometry as described in the Materials and Methods section. The schematic displays the 16 cleavage sites that were identified in SNX1; the previously identified cleavage site is indicated in green. See Supplementary Figure 1 for peptide listing and MS data used to identify these cleavage sites. Proteins were analyzed by immunoblotting using the indicated antibodies. Closed and open arrowheads indicate full-length and cleaved fragments, respectively. Actin was used as a loading control.