Summary
Bacterial wilt caused by Ralstonia solanacearum is a ruinous soilborne disease affecting more than 450 plant species. Efficient control methods for this disease remain unavailable to date. This study characterized a novel nucleotide‐binding site‐leucine‐rich repeat resistance gene AhRRS5 from peanut, which was up‐regulated in both resistant and susceptible peanut cultivars in response to R. solanacearum. The product of AhRRS5 was localized in the nucleus. Furthermore, treatment with phytohormones such as salicylic acid (SA), abscisic acid (ABA), methyl jasmonate (MeJA) and ethephon (ET) increased the transcript level of AhRRS5 with diverse responses between resistant and susceptible peanuts. Abiotic stresses such as drought and cold conditions also changed AhRRS5 expression. Moreover, transient overexpression induced hypersensitive response in Nicotiana benthamiana. Overexpression of AhRRS5 significantly enhanced the resistance of heterogeneous tobacco to R. solanacearum, with diverse resistance levels in different transgenic lines. Several defence‐responsive marker genes in hypersensitive response, including SA, JA and ET signals, were considerably up‐regulated in the transgenic lines as compared with the wild type inoculated with R. solanacearum. Nonexpressor of pathogenesis‐related gene 1 (NPR1) and non‐race‐specific disease resistance 1 were also up‐regulated in response to the pathogen. These results indicate that AhRRS5 participates in the defence response to R. solanacearum through the crosstalk of multiple signalling pathways and the involvement of NPR1 and R gene signals for its resistance. This study may guide the resistance enhancement of peanut and other economic crops to bacterial wilt disease.
Keywords: Arachis hypogaea, resistance gene, bacterial wilt, signal transduction, NPR1, tobacco
Introduction
Bacterial wilt caused by Ralstonia solanacearum is a destructive soilborne bacterial disease in plants, including peanut (Arachis hypogaea L.), worldwide (Wicker et al., 2007). This disease is the key limiting factor for the production yield and quality of peanut, an important oil and food crop in China and the world (Yu et al., 2011). R. solanacearum infects more than 450 plant species, including many important crops, such as peanut, tomato, tobacco, potato, pepper, soybean and rape. However, effective techniques to control this disease remain unavailable to date (Gururani et al., 2012; Yu et al., 2011). The employment of resistant cultivars has been the most efficient strategy to control this disease, but the enhancement has not been conducted successfully in crops thus far (Bhatnagar‐Mathur et al., 2015; Keneni et al., 2012; Reddy, 2016; Sunkara et al., 2014). A recent report has indicated that stable resistant varieties of peanut have been bred to overcome the incidence of serious bacterial wilt in large areas effectively. This report implies that peanut might contain resistant gene resources that are potentially important in controlling this disease. However, few resistant varieties of peanut have been developed in high yield and quality so far (Sunkara et al., 2014). Therefore, elucidating the molecular mechanism underlying the resistance of crops to bacterial wilt is urgently required to breed ideal varieties.
Plants have developed a complete defence mechanism against the infection of pathogens, such as bacteria, viruses, fungi and insects during evolution (Henry et al., 2013; Jones and Dangl, 2006; Thomma et al., 2011; Zvereva and Pooggin, 2012). Several pathogens are killed by the first defence system, whereas some are suppressed by the plant innate immune (PTI) system (Jones and Dangl, 2006; Zhang and Zhou, 2010). Notwithstanding, various successful pathogens deploy effectors for pathogen virulence. Many effectors can interfere with PTI to some extent as effector‐triggered susceptibility (Jones and Dangl, 2006). A given effector is ‘specifically recognized’ by plant NB‐LRR proteins (R genes) during effector‐triggered immunity (ETI) (Jones and Dangl, 2006). In general, R gene‐triggered resistance is associated with a rapid defence response termed hypersensitive response (HR) (Dangl et al., 1996; Greenberg, 1997; Keen, 1990; Thomma et al., 2011). HR brings a localized cell and tissue death at the infection site following a series of downstream defence responses (Baker et al., 1997; Lamb et al., 1989; Ryals et al., 1996; Zvereva and Pooggin, 2012).
NBS‐LRR genes are classified into two subfamilies, namely TIR‐NBS‐LRR and non‐TIR‐NBS‐LRR, on the basis of the motifs located in the N‐terminal region (Liu et al., 2007). The former subfamily contains a Drosophila Toll/mammalian interleukin‐1 receptor (TIR) domain, whereas the latter subfamily consists of a coiled coil (CC)/leucine zip motif (Van Ooijen et al., 2008). Thus far, more than 70 disease resistance genes have been cloned and characterized in monocots and dicots (Liu et al., 2007). Most of these genes are NBS‐LRR genes obtained using map‐based cloning and transposon tagging methods in crops (Hulbert et al., 2001; McDowell and Woffenden, 2003; Meyers et al., 2005; Takken and Joosten, 2000).
R gene products can directly or indirectly recognize pathogen effector proteins (avirulence protein) and induce resistance (Cesari et al., 2013; Flor, 1971; Sohn et al., 2014). Furthermore, some NB‐LRR proteins act downstream of R protein activation. The tobacco ‘N‐required gene 1’ and tomato ‘NB‐LRR protein required for HR‐associated cell death 1’ (NRC1) (both as CC‐NB‐LRR proteins) are required for TIR‐NB‐LRR protein N‐mediated resistance to tobacco mosaic virus and receptor‐like protein Cf‐4‐mediated resistance to tomato leaf mould, respectively (Gabriëls et al., 2007; Peart et al., 2005). The CC‐NB‐LRR activated disease resistance 1 family of proteins in Arabidopsis is required for salicylic acid (SA)‐dependent ETI (Bonardi et al., 2011). The downy mildew resistance locus RPP2 in Arabidopsis Col‐0 comprises two closely linked NB‐LRR genes, RPP2A and RPP2B, for resistance (Sinapidou et al., 2004). The rice Pia locus for blast (Magnaporthe) resistance includes two divergently transcribed CC‐NB‐LRR genes, RGA4 and RGA5, for resistance (Cesari et al., 2013).
Quantitative trait loci (QTL) controlling resistance to bacterial wilt have been identified in several crops, such as tomato (Carmeille et al., 2006; Danesh et al., 1994; Mangin et al., 1999; Thoquet et al., 1996; Wang et al., 2000), eggplant (Lebeau et al., 2013) and tobacco (Qian et al., 2012), as well as in model plants, such as Arabidopsis thaliana (Godiard et al., 2003) and Medicago truncatula (Ben et al., 2013). However, only two resistance genes have been identified thus far: the A. thaliana ERECTA gene involved in polygenic resistance and the A. thaliana RRS1‐R gene involved in monogenic resistance. RRS1‐R is a typical TIR‐NB‐LRR resistance gene generated through map‐based cloning in Arabidopsis (Deslandes et al., 2002). RRS1‐R contains a WRKY transcription factor domain at the C‐terminus to activate downstream gene expression and a nuclear localization signal (NLS) at its N‐terminus (Deslandes et al., 2002). PopP2 is the corresponding avirulence gene of RRS1‐R. It was recognized and recruited with the LRR domain of RRS1‐R and trafficked to the nucleus through NLS. ERECTA, a quantitative resistance locus for bacterial wilt, encodes a leucine‐rich repeat receptor‐like kinase. ERECTA‐controlled resistance is triggered by disease defence response through the phosphorylation of extracellular kinase‐regulated downstream genes (Godiard et al., 2003). However, resistance genes to bacterial wilt have yet to be cloned in crops other than Arabidopsis, thereby hindering genetic enhancement towards the disease. In addition, the molecular mechanism and details in the signalling pathway of R gene resistance to R. solanacearum have yet to be elucidated.
In this study, the up‐regulated NBS‐LRR resistant gene AhRRS5 was screened from peanut through microarray analysis. This gene was induced by R. solanacearum containing the typically conserved motifs of an NBS‐LRR gene. AhRRS5 was localized in the nucleus and could be up‐regulated relatively higher in the resistant than susceptible peanut cultivars against bacterial wilt. This gene responded differently to phytohormones, such as salicylic acid (SA), abscisic acid (ABA), methyl jasmonate (JA) and ethephon (ET), among distinct resistance varieties. The transient overexpression of AhRRS5 induced HR responses in Nicotiana benthamiana, whereas the overexpression of this gene in Nicotiana tabacum significantly enhanced the resistance of peanut to R. solanacearum. The underlying mechanism presumably involved the significant up‐regulation of several representative stress‐responsive and resistance marker genes. We concluded that AhRRS5 indirectly participates in the defence response to R. solanacearum in plants through multiple signalling regulatory networks.
Results
Cloning and phylogenetic analysis of AhRRS5
The 5′ and 3′ unknown cDNA sequences of AhRRS5 were cloned by rapid amplification of cDNA ends (RACE) on the basis of the known fragment. The full‐length cDNA sequence of AhRRS5 was isolated from the total RNA of peanut leaf through reverse transcription polymerase chain reaction (RT‐PCR), and the genomic DNA sequence of AhRRS5 was cloned from the genomic DNA of peanut through PCR. The full‐length cDNA contained a 3157‐bp open reading frame encoding a polypeptide of 943 amino acids, an 88‐bp 5′ untranslated terminal region (5′ UTR), and a 138‐bp 3′ UTR. The genomic DNA sequence of AhRRS5 was 3662‐bp, including a 535‐bp intron. The entire sequence of the AhRRS5 protein has 76% identity with an NBS‐LRR resistance protein, RPM1‐like, in Glycine max (Figure 1; Data S1 and Data S2). A comparison of the AhRRS5 amino acid sequence with the R gene of a known function demonstrates that it most closely resembles RXO1 (33% identity and 53% positive) from Zea mays, which confers resistance to X. o. pv. Oryzicola containing avrRxo1, and RPM1 (32% identity and 53% positive) from A. thaliana, resisting Pseudomonas syringae pv. maculicola 1 containing AvrBand, AvrRpm1, and Pid3 (33% identity and 53% positive) from Rice and resisting Magnaporthe oryzae (Data S2). The former two were resistant to bacterial pathogens.
Figure 1.

Conserved domain comparison between the deduced amino acid sequence of AhRRS5 and other resistance proteins. Sequences were aligned using the ClustalW2 program. Gaps have been introduced to optimize the alignment. Identical or conserved amino acids are shaded in dark and light, respectively. The sources of the proteins and GenBank accession numbers are as follows: OsPid3, blast resistance protein (ACN62383.1) from Oryza sativa Indica Group; AtRPM1 (AGC12570.1) from Arabidopsis thaliana; GmRPM1 (XP_006587620.1) from Glycine max; and ZmRXO1 disease resistance protein (AAX31149.1) from Zea mays.
Sequence analysis showed that the deduced AhRRS5 protein contained conserved NBS motifs, such as P‐loop (MGGVGKT), GLPL (GLPLALK), kinase‐2 (LLVLDDVVW), kinase‐3a (GSRVLVTTR) and RNBS‐C (YEVxxLSDEEAWELFCKxAF) motif (Bertioli et al., 2003; Zheng et al., 2012), and 4 LRR‐conserved domains (LxxLxxLxxLxLxxC/A‐xx) (Leah McHale et al., 2006) (Figure 1; Data S1). On the basis of the conserved domains at the N‐terminus of the deduced NBS‐LRR genes, the AhRRS5 gene had the typical structure of non‐TIR‐NBS‐LRR genes (Wan et al., 2012), with RNBS‐A‐non‐TIR (FnLxAWVCvSQxF) domains (Figure 1).
The phylogenetic analysis of 29 types of NBS‐LRR resistance proteins from GenBank together with AhRRS5 generated two clades coarsely (Figure 2; Data S3). The topology of the phylogenetic analysis showed that the NBS‐LRR‐type resistance proteins can be divided into two types, namely TIR‐NBS‐LRR and non‐TIR‐NBS‐LRR, and that the non‐TIR‐NBS‐LRR‐type resistance proteins can be subdivided into two classes, namely NBS‐LRR and CC‐NBS‐LRR. AhRRS5 is a NBS‐LRR‐type resistance protein that is similar to NBS‐LRR resistance proteins, such as RPM1 (XP_006587620.1|) from Glycine max, RPP8 (GenBank: XP_003612691.1) from M. truncatula, RXO1 (GenBank: AAX31149.1) from Zea mays, RPM1 (GenBank: AGC12590) from A. thaliana and Pi9 (GenBank: ABB88855.1) from Oryza sativa. These similarities indicate that these resistance genes share a common ancestor R gene and belong to NBS‐LRR‐type resistance genes (Figure 2).
Figure 2.
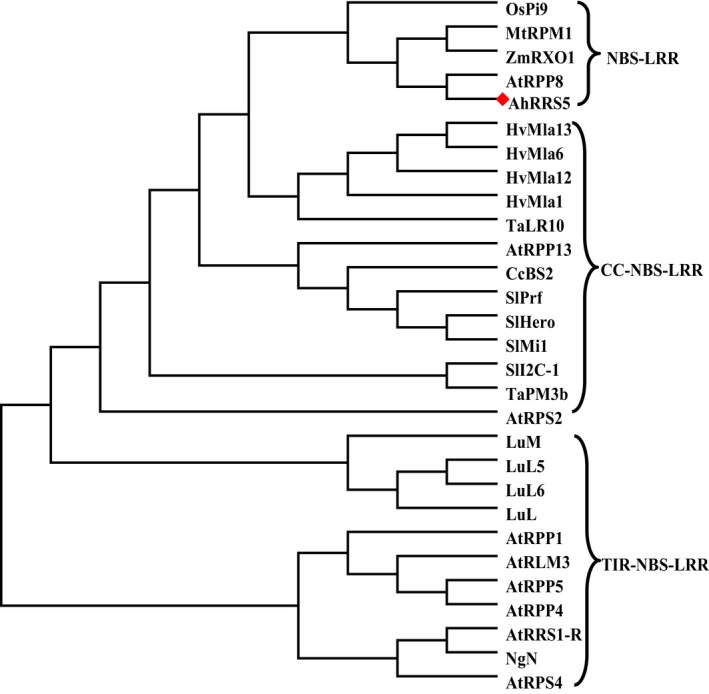
Phylogenetic tree was constructed using AhRRS5 and known different types of NBS‐LRR resistant proteins. AhRRS5 is shown by a red rhombus. Alignments were performed in ClustalW2, and phylogenetic tree was constructed by the neighbour‐joining algorithm of MEGA 5.01. Bootstrap values (1000 replicates) are shown in percentages at the branch nodes.
AhRRS5 functions in the nucleus
Sequence analysis indicated that the predicted AhRRS5 protein was localized in the nucleus (Data S1) (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). To confirm this indication and the site of function, we generated an AhRRS5‐green fluorescent protein (GFP) fusion driven by the constitutive CaMV35S promoter (Figure 3a). With 35S::GFP as a negative control, the AhRRS5::GFP fusion gene was transformed into Agrobacterium strain GV3101, which was further infiltrated into N. benthamiana leaves. Typical results indicated the exclusive localization of AhRRS5‐GFP in the nucleus, whereas GFP alone occurred in multiple subcellular compartments, including the cytoplasm and the nucleus (Figure 3b). The results indicate that AhRRS5 is localized and functions in the nucleus.
Figure 3.
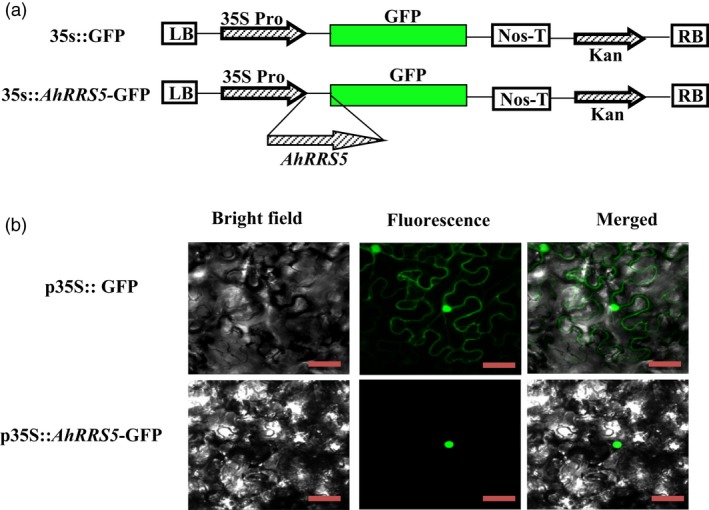
Subcellular localization of AhRRS5. (a) Schematic of p35S::GFP and p35S:: AhRRS5‐GFP constructs used for the subcellular localization of AhRRS5 by agroinfiltration into N. benthamiana cells. (b) AhRRS5‐GFP localized in the nucleus of N. benthamiana cells, GFP alone localized throughout the whole cells. Bright field (left), fluorescence (middle) and merged images (right) were obtained at 48 h by using Leica confocal microscopy after agroinfiltration. Bars = 50 μm.
AhRRS5 showed varied expression patterns among tissues
In the microarray with a high density of unigenes, four unigenes including AhRRS5 were found with a sequence identity of more than 97%. These unigenes apparently belong to the same AhRRS5 gene family. Nonamplified double strain cDNA was used for microarray hybridization to evaluate the transcript levels of the unigenes. All four members showed a synchronized expression pattern among tissues or organs. They showed tissue‐specific expression manners; in particular, they were expressed the highest in the roots, then in the testa, pericarps and stem, but were weakly expressed in other tissues (Figure 4a). Embryos displayed the least expression levels of these genes. In addition, the transcripts of these genes obviously increased with pericarp development (Figure 4b) but remained almost constant with trace amounts during embryo development (Figure 4c). Therefore, AhRRS5 may be involved in the resistant response and in plant development to some extent.
Figure 4.
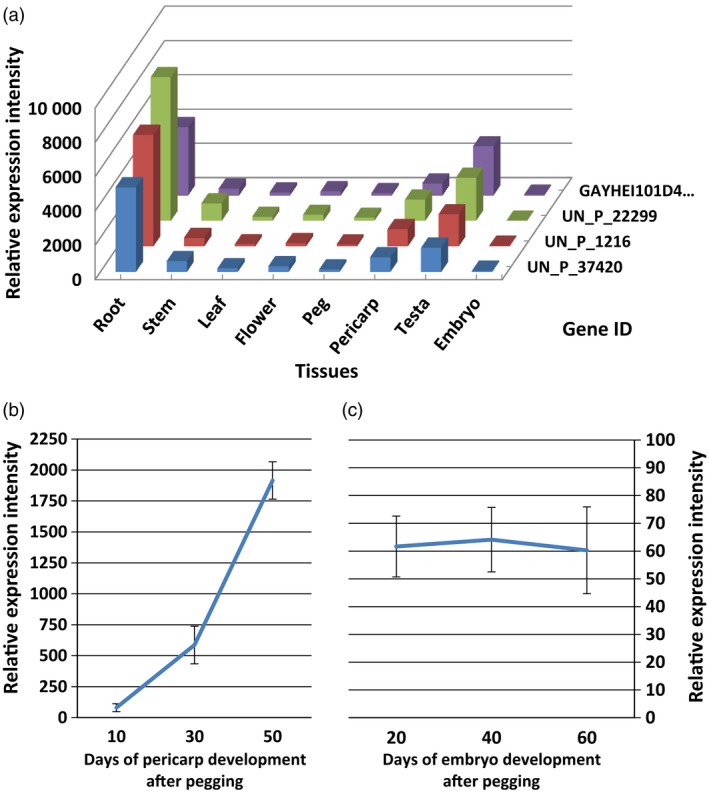
In silico identification of the expression characteristics of four members of the AhRRS5 gene family. (a) The AhRRS5 family showed tissue‐specific expression in peanut, the highest level was in the root, followed by the testa and pericarp. Weak expression was found in the other tissues. (b) AhRRS5 genes increased expression with pericarp development. (c) AhRRS5 had the least expression levels with developing embryos. UN_P_37420, UN_P_1216, UN_P_22299 and GAYHEI101D4L7C_pchu_p are AhRRS5 and the three other members of the same family.
AhRRS5 showed a wide response to biotic and abiotic stresses
Response of AhRRS5 to exogenous hormones
The transcript level of AhRRS5 was determined in the medium‐resistant variety Minhua 6 at the eight‐leaf stage after exogenous treatment with SA, ABA, ET and MeJA to identify the possible involvement of AhRRS5 in signalling pathways relating to the phytohormones (Figure 5). Compared with the control plants, AhRRS5 transcripts increased between 3 and 24 h with two peaks after SA treatment. The highest transcript level (6.7‐fold up‐regulation) was observed at 12 h post‐treatment (hpt) (Figure 5a). AhRRS5 transcription also increased with a single peak of 5.1‐fold up‐regulation at 3 hpt after ABA treatment (Figure 5b). In response to ET, AhRRS5 expression was enhanced from 3 hpt to 12 hpt, and the highest transcript level (10‐fold) was obtained at 12 hpt (Figure 5c). The application of 100 mM MeJA also elevated AhRRS5 expression with two peaks, and the highest level was achieved at 6 hpt (Figure 5d).
Figure 5.
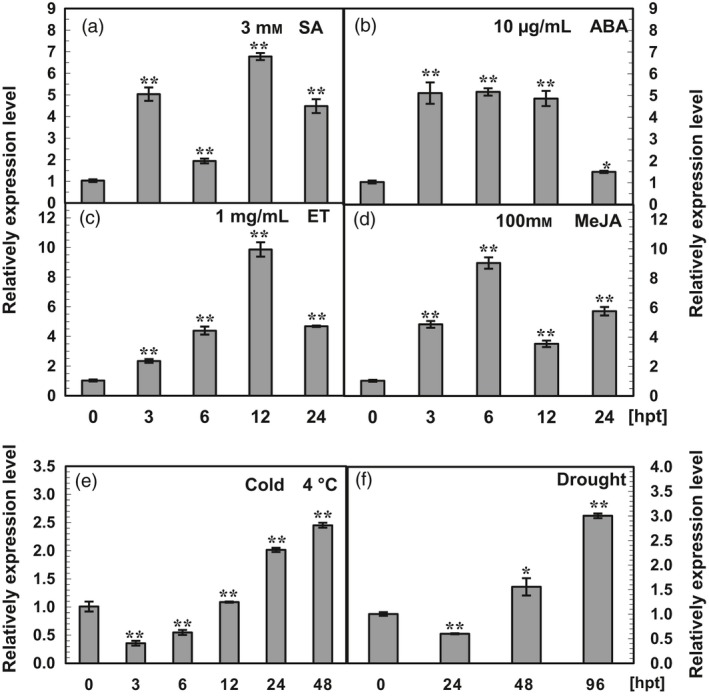
qRT‐PCR analysis of AhRRS5 transcripts in peanut cultivar Minhua 6 under abiotic treatments. (a–d) AhRRS5 relative expression level in peanut leaves at different time points after treatment with salicylic acid (SA, 3 mM), abscisic acid (ABA, 10 μg/mL), ethylene (ET, 1 mg/mL) and methyl jasmonate (MeJA, 100 mM). (e and f) AhRRS5 expression was determined at various hour intervals after treatment with low temperature (4 °C) and drought in peanut plants at the eight‐leaf stage. The relative expression level of AhRRS5 in peanut plants at various time points was compared with the mock control, which was set to 1. The asterisks indicate a significant difference (SNK test, *P < 0.05 or **P < 0.01). Error bars indicate the standard error.
Highly susceptible and resistant varieties Xinhuixiaoli and Yueyou 92, respectively, were used to clarify the relationship between AhRRS5 and the hormones (Figure 6). Although AhRRS5 showed a similar expression in response to these hormones in Minhua 6, this gene demonstrated distinct expression characteristics between the two varieties. AhRRS5 was more significantly up‐regulated after SA and ABA treatments in Xinhuixiaoli than in Yueyou 92 (Figure 6a,b); however, this gene increased less after ET and JA treatments (Figure 6c,d). In particular, the application of ET down‐regulated AhRRS5 in Xinhuixiaoli but up‐regulated it in Yueyou 92 (Figure 6d). This result indicates that the regulation of AhRRS5 differs between resistant and susceptible varieties in peanut.
Figure 6.
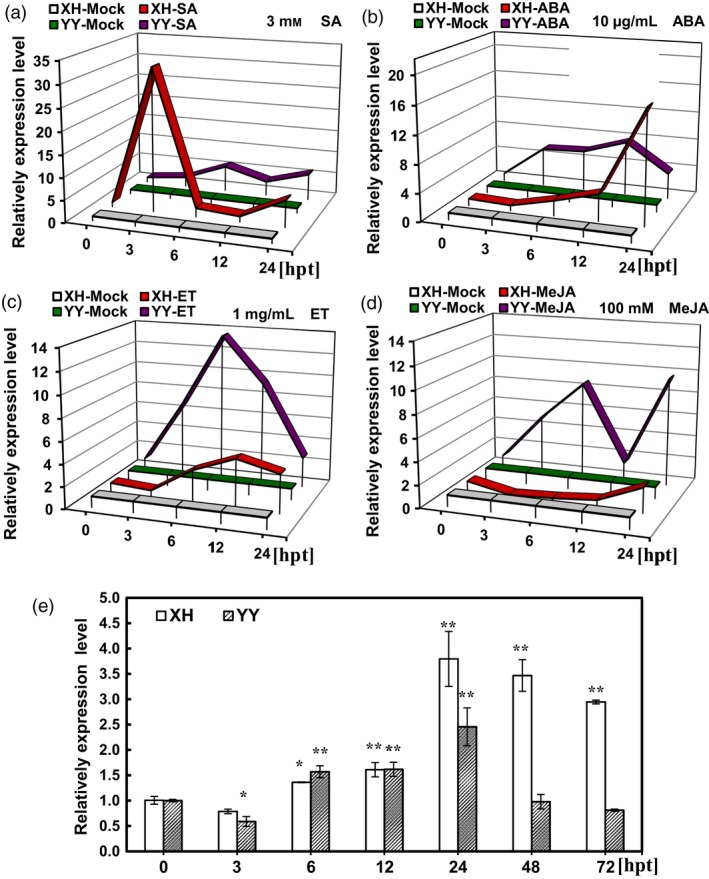
Comparative expression characteristics of AhRRS5 between resistant and susceptible varieties under hormones and R. solanacearum treatments. (a) AhRRS5 showed two expression peaks in response to SA within 24 h, and it up‐regulated over 32‐fold in susceptible variety at 3 HPT, much greater than in resistant one. (b) AhRRS5 increased expression under ABA treatment with one peak; it up‐regulated later in the susceptible but >16‐fold at 24 h. (c) AhRRS5 up‐regulated with one peak within 24 h with nearly 16‐fold at 6 h in the resistant variety. (d) AhRRS5 responded differently between resistant and susceptible peanut with MeJA treatment, down‐regulated in the susceptible peanut and up‐regulated in the resistant ones with two peaks of over eightfold increase. (e) AhRRS5 was up‐regulated higher in susceptible variety especially after 24 hpt with inoculation of R. solanacearum. XH‐Mock: susceptible variety Xinhuixiaoli without treatment; YY‐Mock: resistant variety Yueyou 92 without treatment. XH‐SA, XH‐ABA, XH‐ET and XH‐MeJA: susceptible variety treated with SA, ABA, ET and MeJA, respectively; YY‐SA, YY‐ABA, YY‐ET and YY‐MJA, resistant variety treated with SA, ABA, ET and MeJA, respectively. The relative expression level of AhRRS5 in peanut plants at various time course was compared with mock or control, which was set to 1. The asterisk indicate a significant difference (SNK test,*P‐value <0.05 or **P‐value <0.01), Error bars indicate the standard error.
Responses of AhRRS5 transcripts to abiotic stresses
The responses of AhRRS5 including three other orthologous NBS‐LRR genes to low temperature (4 °C) and drought were studied by microarray hybridization using the cDNA of mixed double strains at different time points (Materials and methods) in eight‐leaf Minhua 6. AhRRS5 and three other NBS‐LRR genes remained constant in response to low temperature but were up‐regulated by nearly 8‐ to 10‐fold in response to drought (Data S4). To clarify whether AhRRS5 is involved in the response to abiotic stresses, the relative transcripts of AhRRS5 were also examined in eight‐leaf Minhua 6 seedlings under low temperature and drought treatments through quantitative real‐time PCR analysis (Figure 5e,f). The transcript level of AhRRS5 decreased and then increased in response to low temperature and drought. In specific, under low temperature, the transcript level of AhRRS5 decreased by two‐ to three‐fold at 3 and 6 hpt and then increased between 24 and 48 hpt, with the highest level (2.5‐fold) at 48 hpt (Figure 5e). Compared with the control, the transcript level of AhRRS5 was down‐regulated by two‐fold at 1 day post‐treatment (dpt) but was up‐regulated from 2 dpt to 4 dpt with a 3.3‐fold induction at 4 dpt under drought (Figure 5f), thereby confirming the microarray results.
Expression pattern of AhRRS5 in the resistant/susceptible peanut cultivars after R. solanacearum challenge
AhRRS5 was characterized using resistant and susceptible peanut cultivars after inoculation with R. solanacearum by microarray hybridization and qRT‐PCR. The four members of AhRRS5 in the microarray exhibited similar pattern of transcription with R. solanacearum inoculation. These genes were up‐regulated by nearly one‐fold under inoculation with R. solanacearum in Yueyou 92, but a higher up‐regulation was observed in Xinhuixiaoli (Data S4). In addition, the expression patterns of AhRRS5 at different time courses after R. solanacearum inoculation were compared in the two varieties. AhRRS5 transcripts were induced between 0 and 24 h in the leaves of Yueyou 92 and then returned to their ground state at 72 hpi in response to R. solanacearum strain challenge. The expression level of AhRRS5 in Xinhuixiaoli was up‐regulated from 6 hpi, showed a peak of 3.75‐fold transcript level at 24 hpi, and remained high between 24 and 96 hpi (Figure 6e). This finding suggests that AhRRS5 participates in the immunity of peanut to R. solanacearum.
Transient overexpression of AhRRS5 in N. benthamiana leaves induces hypersensitive response
Successful pathogens can attenuate PTI by secreting effector molecules into the host plant cell. Some R proteins could recognize pathogen effector molecules and induce ETI with HR resulting in cell death at the infection site. This process is followed by a series of downstream defence responses. Overexpression vector harbouring p35S::AhRRS5 was generated and transformed into Agrobacterium GV3101 to verify whether AhRRS5 overexpression causes HR cell death. AhRRS5 was transiently expressed in N. benthamiana leaves through agroinfiltration. Then, AhRRS5 overexpression in N. benthamiana leaves induced an intensive HR mimicking cell death 48 h after infiltration. However, no visible HR cell death was found in those infiltrated with GV3101 harbouring empty vector p35S::00. Furthermore, electrolyte significantly leaked at 24 and 48 hpt after treatment, and darker trypan blue staining was observed after AhRRS5 overexpression for 24 hpt. This result suggests that AhRRS5 can trigger HR response in N. benthamiana leaves (Figure 7a,b). In addition, large amounts of H2O2 accumulation were found in the N. benthamiana leaves after AhRRS5 overexpression by DAB staining (Figure 7b). These results demonstrate that the transient overexpression of AhRRS5 in tobacco leaves induces HR and H2O2 generation as a defence response to stresses.
Figure 7.
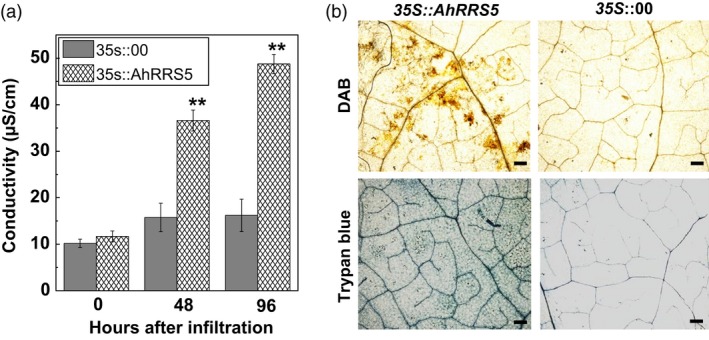
Effect of transient expression of AhRRS5 in Nicotiana benthamiana on immunity induction. (a) Electrolyte leakage of N. benthamiana leaves were infiltrated with Agrobacterium strain GV3101 containing 35S::AhRRS5 and 35S::00. (b) Trypan blue and DAB staining of cell death and H2O2 generation in N. benthamiana leaves 48 h after AhRRS5–Agrobacterium infiltration. Bars = 0.1 mm. Error bars indicate the standard error, Alphabet indicates statistically significant differences between wild‐type and 35S::AhRRS5 tobacco by Student–Newman–Keuls test, *P < 0.05 or **P < 0.01), Error bars indicate the standard error.
Overexpression of AhRRS5 in tobacco enhances resistance to R. solanacearum
The involvement of AhRRS5 R. solanacearum resistance was evaluated by transforming CB‐1, a conventional tobacco cv., medium‐susceptible to bacterial wilt mediated by Agrobacterium, with AhRRS5 driven by two copies of the CaMV35S promoter in the pBI121 binary vector. Transgenic T0 and T1 tobacco plants were generated to examine the role of AhRRS5 in tobacco–R. solanacearum interaction (Figure 8a). Three T2 transgenic homozygous lines were screened by inoculation and identified for their resistance to R. solanacearum (Figure 8b). The line AhRRS5‐OE‐3 line which showed the greatest AhRRS5 relative transcript levels and resistant to R. solanacearum (not shown) of all the tested lines, was selected for the detailed disease resistance assays. No apparent phenotypic differences between the wild‐type and transgenic plants were observed. A highly virulent strain of R. solanacearum was used to inoculate individuals of AhRRS5‐OE‐3 T2 lines and wild‐type plants. Vein injection was then used for R. solanacearum inoculation. All tested transgenic lines exhibited enhanced disease resistance. Evident wilting symptoms were detected in the leaves of wild‐type plants at 7 dpi, whereas only faint wilting symptoms were exhibited by AhRRS5‐OE‐3 lines (Figure 8b,d). Extremely severe wilting symptoms were developed in the wild‐type plants at 20 dpi but not in the AhRRS5‐OE‐3 transgenic lines. Wilting and contagion symptoms were evident on the stems of the infected wild‐type tobacco at 7 and 20 dpi, but no significant symptoms were found in the transgenic lines (Figure 8e). Further evaluation of AhRRS5 was performed in the Honghuadajinyuan cultivar, which is hypersusceptible to R. solanacearum. Five transgenic T2 homozygous lines were inoculated compared with the wild type. All lines showed increased but distinct levels of resistance to R. solanacearum (Tables 1 and S2). Line 3 showed the highest resistance with a low infection index and a death rate of (7.08%) at 21 dpi, but the mock line showed serious wilting with 93.58% index and 81.08% death of plants, respectively, at 21 dpi. These results indicate that AhRRS5 overexpression greatly enhances disease resistance against R. solanacearum in tobacco.
Figure 8.
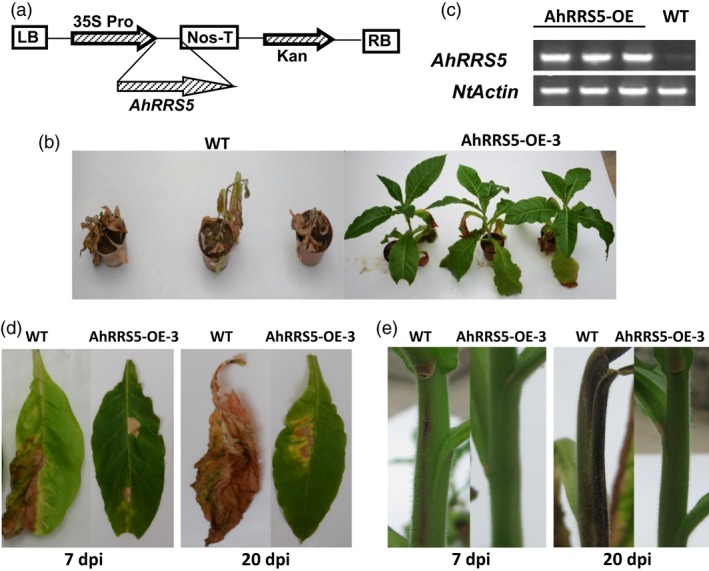
Overexpression of AhRRS5 enhanced resistance to Ralstonia solanacearum in transgenic tobacco. (a) Schematic of the pBI121‐AhRRS5 construct. LB and RB, left and right borders of the T‐DNA; 2 × 35SPro, two cauliflower mosaic virus 35S promoters; Nos‐T, NOS terminator; Kanr, kanamycin resistance. (b) The third leaves of 8‐week‐old wild‐type tobacco and AhRRS5‐OE‐3 transgenic plants were inoculated with 10 μL of suspension of 108 cfu per millilitre of high‐virulence R. solanacearum strain. The photograph was obtained 20 days postinoculation (dpi). (c) RT‐PCR analysis of AhRRS5 expression in transgenic and wild‐type tobacco plants; expression level of Ntactin was visualized as endogenous control. (d) Disease symptoms of detached leaves of wild‐type and AhRRS5‐OE‐3 transgenic plants after inoculation with R. solanacearum. Transgenic leaves showed immune resistance or high‐resistance phenotype. Photos were obtained at 7 and 20 dpi. (e) Different phenotypes of the stem were observed between wild‐type and transgenic AhRRS5‐OE‐3 plants after inoculation with R. solanacearum. Transgenic plant stem showed no or much week infections. Photos were taken at 7 and 20 dpi.
Table 1.
Disease indexes and death ratios of different OE lines and the wild type after inoculation with Ralstonia solanacearum
| OE lines | 7 dpi | 21 dpi | ||
|---|---|---|---|---|
| Disease index (%) | Death ratio (%) | Disease index (%) | Death ratio (%) | |
| OE‐2 | 22.90 | 2.80 | 45.79 | 34.58 |
| OE‐3 | 12.83 | 0.00 | 20.35 | 7.08 |
| OE‐4 | 26.51 | 4.82 | 64.46 | 56.63 |
| OE‐5 | 37.39 | 10.62 | 72.35 | 60.18 |
| OE‐8 | 19.92 | 6.50 | 31.10 | 14.63 |
| Wild type | 73.65 | 22.97 | 93.58 | 81.08 |
dpi, days postinoculation.
To further confirm the role of AhRRS5 in disease resistance and elucidate its possible molecular mode of action, transcriptional responses of known defence genes to overexpression of AhRRS5 in noninoculated tobacco plants were investigated by qPCR (Data S5). We examined transcript levels of the HR‐associated genes NtHIN1, NtHSR201, NtHSR203 and NtHSR515 (Sohn et al., 2007), SA‐responsive genes NtPR1a/c, NtPR3, NtPR4 and NtNPR1 (Brogue et al., 1991; Ward et al., 1991), JA‐responsive NtPR1b and NtPR2 (Sohn et al., 2007) and ET‐associated genes such as NtEFE26 and NtACS6 (Chen et al., 2003). Each of the tested tobacco genes was shown previously to be up‐regulated in response to pathogen infection (Chen et al., 2003; Rizhsky et al., 2002; Sohn et al., 2007). We found transcript levels of HR‐associated genes, such as NtHIN1, NtHSR201 and NtHSR515 to be increased by 3.3‐fold, 2.8‐fold and 3.3‐fold in the AhRRS5‐OE‐3 line compared to wild‐type plants, respectively. Transcript levels of the SA‐responsive NtPR1a/c, NtPR3, NtPR4 and NtNPR1 genes were increased in AhRRS5‐OE‐3 plants by 11.9‐fold, 3.0‐fold, 2.0‐fold and 3.0‐fold, respectively, while those of the JA‐responsive NtPR2 and NtPR1b genes were 2.5‐fold and 4.0‐fold higher in AhRRS5‐OE‐3 plants. These results show that AhRRS5 overexpression enhances stress‐related gene expression compared to the wild‐type tobacco.
Up‐regulation of marker genes in response to R. solanacearum infection
HR‐responsive genes, namely NtHIN1, NtHSR201 and NtHSR515, were significantly up‐regulated in the transgenic plants (P < 0.01 or P < 0.05) but down‐regulated in wild‐type CB‐1 to different extents at 48 hpi with R. solanacearum (Figure 9). By contrast, NtHSR203 did not respond to the strain infection either in the transgenic or control plants (Figure 9a). The expression levels of NtPR1a/c and NtPR3, which are SA‐responsive pathogenesis‐related (PR) genes, increased in the AhRRS5‐OE‐1 plants by 1, 453.0‐ and 14.5‐fold, respectively, which are much higher than those in CB‐1. In addition, the NtRP4 gene was down‐regulated by 2.5‐fold (Figure 9b). JA‐responsive NtPR2 was up‐regulated in CB‐1 but down‐regulated in the transgenic plants in response to the strain, whereas NtPR1b was 14.3‐fold higher in the AhRRS5‐OE‐3 plants than in CB‐1 (Figure 9c). The transcript levels of ET‐responsive genes NtEFE26 and NtACS6 in the transgenic plants were also significantly increased at 48 h after infection but not in the wild‐type plants (Figure 9d). Several pathogen‐induced HR‐ and defence‐associated genes were enhanced by AhRRS5 overexpression, but few were reduced or remained unchanged, which are consistent with the resistance enhancement in the transgenic lines. These findings indicate that AhRRS5 functions in the resistance of transgenic tobacco through a wide series of signalling pathways.
Figure 9.
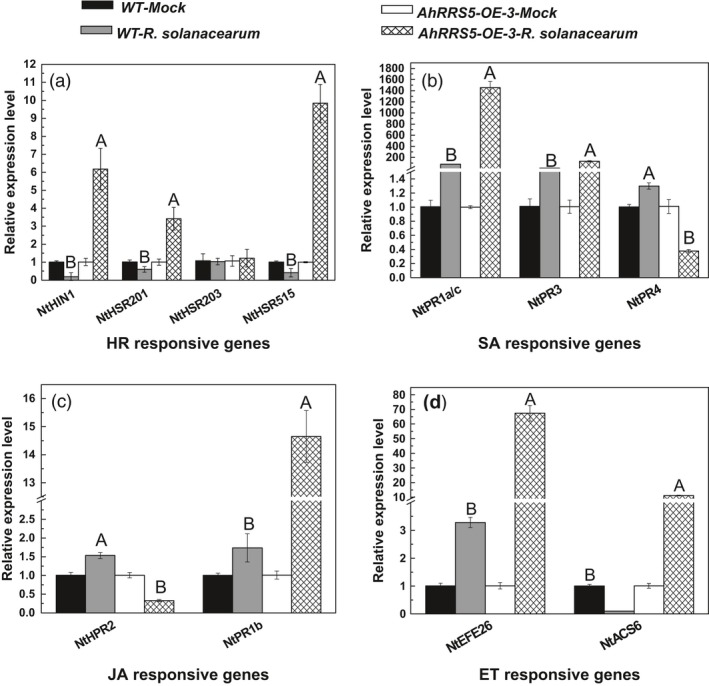
Transcript levels of tobacco defence‐related marker genes in wild‐type CB‐1 and AhRRS5‐OE‐3 transgenic tobacco line 48 h after inoculation with R. solanacearum. The transcript levels of NtHIN1, NtHSR201, NtHSR203, NtHSR515, NtPR1a/c, NtPR3, NtPR4, NtNPR1, NtPR2, NtPR1b, NtEFE26 and NtACS6 were determined by quantitative real‐time PCR. Relative transcript levels were normalized using the transcripts of NtEF1α. The transcript levels of nontreated wild‐type or AhRRS5‐OE‐3 tobacco plants were used as the control and assigned value of 1. Alphabet indicates statistically significant differences between wild‐type and AhRRS5‐OE‐3 tobacco plants by Student–Newman–Keuls test (lowercase difference indicates P < 0.05; uppercase difference indicates P < 0.01).
NDR1 and NPR1 genes were up‐regulated by R. solanacearum infection
Non‐race‐specific disease resistance 1 (NDR1) and nonexpressor of pathogenesis‐related gene 1 (NPR1) genes were involved in the R gene resistance signalling pathway. In silico identification of three NDR1‐like and two NPR1‐like gene expressions were performed between AhRRS5‐OE‐3 transgenic plants and wild‐type plants, as well as hyper‐resistant and hypersusceptible varieties Yanyan 97 and Honghuadajinyuan after inoculation with R. solanacearum, respectively (Figure 10). Two NPR1‐like genes were slightly up‐regulated by 6%–23% in the AhRRS5‐OE‐3 lines after inoculation but were down‐regulated by 15%–21% in the wild‐type plants after inoculation (Figure 10a). The NPR1 gene, TC79797, considerably increased or decreased in response to the pathogen, consistent with the resistant and susceptible varieties after inoculation (Figure 10b). Furthermore, the results of real‐time PCR revealed that the transcript level of NPR1 increased by 14.5‐fold in transgenic lines of AhRRS5‐OE‐3 as compared with wild‐type plants after inoculation with R. solanacearum, much higher than the increase of transcripts in inoculated wild‐type over corresponding mock plants (Figure 10c).
Figure 10.
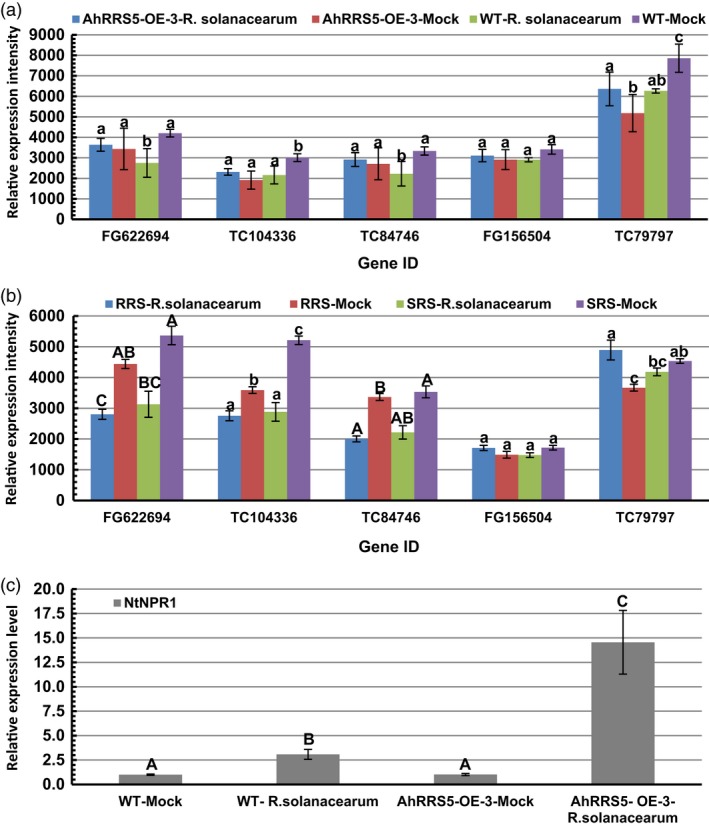
In silico and qPCR analysis of NDR1‐ and NPR1‐like gene expression upon inoculation with R. solanacearum. (a and b) Microarray data. (a) Expression of three NDR1‐like and two NPR1‐like genes. AhRRS5‐OE‐3‐R. solanacearum indicates tobacco CB‐1 cultivar transformed with AhRRS5 with inoculation; AhRRS5‐OE‐3‐Mock, transgenic CB‐1 without inoculation; WT‐R. solanacearum, CB‐1 with inoculation; WT‐Mock, CB‐1 without inoculation. (b) Down‐regulation of three NDR1‐like genes in varieties after inoculation. RRS ‐R. solanacearum indicates hyper‐resistant tobacco variety Yanyan 97 under inoculation; RRS‐Mock, hyper‐resistant variety Yanyan 97 without inoculation. SRS R. solanacearum, hypersusceptible variety Honghuadajinyuan with inoculation; SRS‐Mock, hypersusceptible variety Honghuadajinyuan without inoculation. FG622694,TC104336 and TC84746 are NDR1‐like genes; FG156504 and TC79797 are NPR1/NIM1‐like genes, respectively. (c) Transcript level of NtNPR1 gene in tobacco plants with or without inoculation with R. solanacearum through qRT‐PCR analysis. WT‐Mock and WT‐R. solanacearum, AhRRS5‐OE‐3‐Mock and AhRRS5‐OE‐3‐R. solanacearum indicate wild‐type tobacco without or with inoculation with pathogen, AhRRS5‐OE‐3 transgenic tobacco without or with inoculation with pathogen, respectively. Alphabets mark statistically significant differences between wild‐type and transgenic tobacco plants, by Student–Newman–Keuls test (lowercase differences indicate P‐value <0.05; uppercase differences indicate P‐value <0.01).
The transcript levels of three NDR1‐like genes slightly increased in the AhRRS5 transgenic lines but significantly decreased in the wild type after inoculation; this result indicates that AhRRS5 can maintain a high level of expression for the NDR1 gene (Figure 10a). However, three NDR1‐like genes were considerably down‐regulated in both resistant and susceptible varieties after inoculation (Figure 10b). The results indicate that both NDR1‐ and NPR1‐like genes in tobacco are involved in AhRRS5 resistance in transgenic tobacco, but only NPR1 genes are required for the hyper‐resistant tobacco variety Yanyan 97. AhRRS5 might also be involved in the R gene signalling for resistance against microbial infection.
Discussion
AhRRS5 is a novel peanut NBS‐LRR resistance protein localized in the nucleus
NBS‐LRR genes are a class of resistance genes that function in pathogen recognition and defence response signal transduction (Ameline‐Torregrosa et al., 2008; Gao et al., 2010). More than 70 disease resistance genes cloned from higher plants by map‐based methods belong to NBS‐LRR domain genes resistant to bacterial, fungal and viral diseases, as well as some environmental stresses (Liu et al., 2007). AhRRS5 was isolated from peanut using microarray analysis and could be up‐regulated by R. solanacearum inoculation. The AhRRS5 protein has a typical NB‐ARC domain containing P‐loop, kinase‐2, kinase‐3a and GLPL and other conservative modules similar to Arabidopsis RPM1, RXO1 protein of maize, Pid3 of rice and so on (Figure 1; Leister et al.,1996; Zhao et al., 2004; Chen et al., 2011). Four normal LRR motifs, which may participate in the peanut pathogen interaction or defence responses against the pathogen, were found in AhRRS5 (Takken and Joosten, 2000). The revealed amino acid sequence of AhRRS5 most closely resembles those of R genes of known functions, such as RXO1 from Z. mays resistant to Xanthomonas oryzae pv. Oryzicola (Zhao et al., 2004, 2005), RPM1 from A. thaliana resistant to P. syringae (Leister et al., 1996) and Pid3 from rice resistant to M. oryzae (Chen et al., 2011) (Data S2). Phylogenetic analysis with 29 R genes of known functions showed that AhRRS5 could be classified into non‐TIR‐NBS‐LRR type and NBS‐LRR subclass of resistance genes.
Subcellular localization visualized by the AhRRS5::GFP fusion protein in N. benthamiana leave cells showed that the AhRRS5::GFP fusion protein appeared solely in the nucleus and was associated with its nuclear localization signal GKFKKLKILGLDRF at positions 816–829 (Figure 3b; Data S1). This result agrees with the subcellular localization features of most NBS‐LRR disease resistance genes (Meyers et al., 2003). The first identified resistance gene to bacterial wilt is RRS1‐R (a TIR‐NBS‐LRR gene) in Arabidopsis, which is mainly cytoplasm‐localized but nuclear‐localized only depending on the presence of effector PopP2 from R. solanacearum (Deslandes et al., 2003). The RRS1‐R protein contains TIR‐, NBS‐ and LRR‐conserved domains, aside from a WRKY motif, which activates transcription in plants (Eulgem and Somssich, 2007). Another NBS‐LRR resistance gene, RPS4, to R. solanacearum in Arabidopsis is also localized in both the nucleus and cytoplasm (Wirthmueller et al., 2007). However, RPM1 activated in the plasma membrane functions independent of the nucleus (Boyes et al., 1998; Gao et al., 2011). Therefore, AhRRS5 possibly functions mainly in the nucleus.
AhRRS5 is widely involved in defence responses to biotic/abiotic stresses
AhRRS5 transcripts were up‐regulated in both resistant and susceptible varieties challenged with R. solanacearum and highly up‐regulated in the susceptible variety at 24 hpi (Figure 6e). These results indicate that AhRRS5 participates in the defence response to the pathogen. AhRRS5 was up‐regulated in response to all exogenous hormones applied, namely SA, ABA, ET and JA, in the leaves, although this gene was specifically expressed in the peanut root, testa and pericarp, and weakly in other organs, such as the leaf (Figure 4a). These phytohormones are well‐known signalling molecules involved in controlling the defence gene expression against biotic and abiotic stresses (Divi et al., 2010; Ton et al., 2009). SA is usually associated with R gene‐mediated disease resistance, and SA‐deficient mutants often compromise R gene‐mediated resistance (Yang et al., 2013). Exogenous application of SA induces PR genes and enhances resistance to a broad range of pathogens (Bari and Jones, 2009). Arabidopsis RRS1‐R‐mediated resistance to R. solanacearum is partially dependent on SA and NDR1 (Deslandes et al., 2002). Arabidopsis RCY1 gene, which encodes a CC‐NBS‐LRR protein for resistance to the yellow strain of cucumber mosaic virus, requires SA and ET signalling (Takahashi et al., 2002). ET regulates various growth and developmental processes and is also involved in responses to stresses, such as salt, drought, cold, flooding and infection caused by microbes and insects (Yoo et al., 2009). ET could modulate disease resistance (Broekaert et al., 2006; Van Loon et al., 2006). MeJA regulate defence to herbivores and necrotrophic pathogens (Browse, 2009). SA and JA/ET defence pathways are usually antagonistic, but synergistic interactions have also been reported in defence response to pathogens (Beckers and Spoel, 2006; Mur et al., 2006; Nahar et al., 2012; Vos et al., 2015), which is also consistent with the results on AhRRS5 responding to phytohormones such as SA, JA and ET. Rice ET, JA and SA biosynthetic pathways are prerequisites for defence against Hirschmanniella oryzae, and ABA participates in the antagonistic interaction to SA/JA/ET‐dependent basal defence to the pathogen (Nahar et al., 2012). We found AhRRS5 was up‐regulated in response to all of the four hormones including ABA. ABA functions in abiotic stress tolerance, antagonizes the SA signalling pathway in higher plants and enhances disease susceptibility (Bari and Jones, 2009; Jiang et al., 2010; Nahar et al., 2012). However, ABA plays a positive role in papilla‐mediated defence against Leptosphaeria maculans in Arabidopsis (Ton et al., 2009). Exogenous application of ABA strengthens rice basal resistance against the brown spot caused by Cochliobolus miyabeanus (De Vleesschauwer et al., 2012). The role of ABA in defence depends on the type of pathogens, timing of the defence response and plant tissues (Ton et al., 2009). In general, hormone balance plays a vital role in fine tuning appropriate defence responses to the recognized pathogen.
In the present study, the results of qRT‐PCR and microarray analysis showed that AhRRS5 was up‐regulated by SA, ABA, ET and JA and was enhanced differently in the response to R. solanacearum in three resistant varieties. Concentration curves showed that AhRRS5 was up‐regulated with two optimal peaks in response to SA and JA, but with a single peak to ABA and ET in Minhua 6 (Figure 5a–d). Similar patterns were also found in Xinhuixiaoli and Yueyou 92, although AhRRS5 was down‐regulated in Xinhuixiaoli 24 h after JA treatment (Figure 6a–d). These results indicate that AhRRS5 may involve in the crosstalk between these phytohormones against pathogen infection, such as R. solanacearum. AhRRS5 also showed an altered response to low temperature and drought (Figure 5e,f), indicating its association with biotic/abiotic stresses. Our data suggest that peanut AhRRS5 plays a role in the defence response to bacterial wilt via the synergistic interaction of diverse signalling pathways. Therefore, AhRRS5 in response to R. solanacearum may adopt a distant mechanism in comparison with other pathogen‐associated genes.
AhRRS5 confers resistance to bacterial wilt in heterozygous tobacco transformant
The resistance genes against R. solanacearum have not been cloned and characterized except for model plant Arabidopsis (Deslandes et al., 2002; Godiard et al., 2003). AtRRS1‐R, genetically identified as recessive, confers dominant resistance to R. solanacearum GMI1000 in transgenic Arabidopsis. This gene presents a novel R gene structure combining domains of a TIR‐NBS‐LRR protein and a WRKY motif (Deslandes et al., 2002). Deslandes et al. (2003) showed that RRS1 can recognize the pathogen by directly interacting with effector PopP2 and depends on PopP2 to colocalize at the nucleus for pathogen defence. The Arabidopsis LRR‐RLK gene ERECTA, located in the QTL QRS1, shows resistance to R. solanacearum and also affects the development of aerial organs (Godiard et al., 2003). The NB‐LRR gene RPS4 from Arabidopsis ecotype Ws‐0 functions as a dual resistance gene system with RRS1 to prevent three distinct pathogens, namely R. solanacearum, Pst‐avrRps4 and Colletotrichum higginsianum (Narusaka et al., 2009). RPS4 was suggested to function downstream of, or together with, RRS1‐Ws in the signalling pathway resistant to R. solanacearum.
AhRRS5 induced by R. solanacearum challenge is a non‐TIR‐NBS‐LRR gene different from RRS1‐R in Arabidopsis. Overexpression transgenic tobacco constitutively expressing AhRRS5 showed enhanced disease resistance to bacterial wilt. In specific, AhRRS5 overexpression in transgenic CB‐1, a medium‐susceptible cultivar, showed strong resistance to the pathogen infection (Figure 8). The hypersusceptible cultivar Honghuadajinyuan overexpressing AhRRS5 also increased the resistance to R. solanacearum infection, although different transgenic lines demonstrated distinct levels of resistance in response to the pathogen (Table 1). Lines OE‐3 and OE‐8 showed much higher resistance or immune response to bacterial wilt than other lines, which may have resulted from the effect of insertion locations of the gene in chromosomes. The transient overexpression of AhRRS5 in N. benthamiana showed that it can induce hypersensitive response causing cell death and also produce H2O2 in HR (Figure 7). These results indicate that AhRRS5 may participate in resistance against R. solanacearum involving ROS signalling. Therefore, AhRRS5 is a novel NBS‐LRR resistance gene cloned from peanut, which confers resistance to the R. solanacearum.
AhRRS5 resistance is involved in multidefence signalling pathways
A complex network of different signalling transductions exists in plant–pathogen interactions, and different signalling pathways are associated with the transcription of some marker genes in their mediated disease resistance reaction. Many marker genes, such as NtHIN1, HSR201 and HSR515, are activated in HR signalling (Sohn et al., 2007). SA‐mediated defence responses could activate system‐acquired resistance (SAR) and are accompanied with the expression of several PR genes, such as PR1a/c, PR3, PR4 and PR5 (Dong, 1998; Glazebrook, 2005). PR genes PR2 and PR1b are activated and expressed in ET‐mediated defence response, whereas EFE26 and ACS6 are activated in JA‐mediated defence response (Koornneef and Pieterse, 2008; Kunkel and Brooks, 2002; Thomma et al., 1998). Changes in the expression levels of these markers directly indicate the involvement of plant defence responses and signal transduction pathways (Chen et al., 2003; Rizhsky et al., 2002; Sohn et al., 2007). We examined the transcripts of these marker genes in AhRRS5 overexpression tobacco lines by qPCR. Results showed that AhRRS5 overexpression up‐regulated not only the transcript levels of NtHIN1, NtHSR201 and NtHSR515 in HR signalling but also those of SA‐ regulated genes (PR1a/c, PR3) in the T2 tobacco plants inoculated with virulent R. solanacearum (Figure 9a,b). The transcript levels of JA‐regulated PR1b and ET‐responsive NtEFE26 and NtACS6 were also greatly enhanced (Figure 9c,d). The results conform to the data in peanut, in which AhRRS5 was up‐regulated by the exogenous applications of SA, ET, JA and ABA. The RRS1‐R‐mediated bacterial wilt resistance in Arabidopsis involves ABA participation, and the effect of ABA is greater than that of SA (Deslandes et al., 2003; Hernández‐Blanco et al., 2007). These results are relatively similar to AhRRS5 response to R. solanacearum, indicating that these hormone signals perform synergistically against the pathogen. The overexpression of AhRRS5 conferring increased resistance to bacterial wilt in tobacco was achieved by the increase the gene expression in defence signal transduction pathways.
AhRRS5 resistance requires the involvement of NDR1 and NPR1
AhRRS5 overexpression up‐regulated NDR1 transcripts in response to R. solanacearum challenge, concurring with the report that RRS1‐R in Arabidopsis is SA‐dependent and requires the downstream gene NDR1 for its resistance to bacterial wilt (cf., Chen et al., 2003). However, NDR1 was significantly down‐regulated in the nontransgenic resistance variety Yanyan 97 in response to the pathogen. This finding indicates that other resistance mechanisms exist in response to bacterial wilt. NDR1 primarily mediates signalling derived from the CC‐NB‐LRR type of R proteins, whereas EDS1 involves those from the TIR‐NB‐LRR class of R proteins (Aarts et al., 1998; Wang et al., 2014). These results are apparently contradictory to the events of AhRRS5 and AtRRS1‐R (Deslandes et al., 2002; Lahaye, 2002). NDR1 involves R protein‐mediated resistance to many pathogens (Day et al., 2006; Lu et al., 2013; Repetti et al., 2004). Soya bean GmNDR1a and GmNDR1b bind pathogen effectors and regulate resistance signalling (Selote et al., 2014). Arabidopsis resistance signalling pathways to P. syringae 2 and P. syringae pv. maculicola 1 exhibit different mechanisms of activation in terms of effector action, but both require NDR1 participation (Kim, 2006). Thus, AhRRS5 is associated with NDR1 for its mediated resistance to bacterial wilt.
NPR1 is a key regulator of SAR and is essential for the SA signal transduction to activate PR gene expression (Pieterse and Van Loon, 2004; Sandhu et al., 2009). We examined NPR1 transcription by employing microarray analysis and found that the transgenic plants overexpressing AhRRS5 up‐regulated the two NPR1 transcripts after inoculation with R. solanacearum but down‐regulated them after pathogen challenge in wild‐type plants (Figure 10a,b). These results were confirmed in the resistant and susceptible varieties, indicating that NPR1 plays an important role in pathogen resistance. We further found that the AhRRS5‐OE‐3 line significantly up‐regulated the transcript level of NPR1 by 14.5‐fold in response to the R. solanacearum challenge (Figure 10c). The PR marker genes of SA signalling in the transgenic plants of AhRRS5 were then up‐regulated (Figure 9b). NPR1‐mediated signalling resisting viral and bacterial pathogens and repressing NPR1 transcript would increase the susceptibility of plants to pathogens (Li et al., 2012; Xiao and Chye, 2011). Thus, our results suggest that AhRRS5 participates in pathogen resistance by employing the NPR1‐mediated SA signalling and the R gene pathway associated with NDR1.
Experimental procedures
Plant materials and growth conditions
Peanut cultivars (Arachis hypogaea cv. Minhua 6, cv. Yueyou 92 and cv. Xinhuixiaoli, as medium‐resistant, hyper‐resistant and hyper‐susceptible variants to R. solanacearum, respectively) were provided by the Oil Crop Institute in Fujian Agriculture and Forestry University. Seeds were sown in sterile sands in plastic pots. Seedlings of transgenic lines and wild‐type tobacco (Nicotiana tabacum cv. CB‐1, cv. Yanyan 97 and cv. Honghuadajinyuan, with medium susceptibility, hyper‐resistance and hyper‐susceptibility to R. solanacearum, respectively) were provided by Fujian Tobacco Agricultural Research Institute. N. benthamiana is available in this laboratory. T2 seeds of transgenic tobacco lines were surface‐sterilized with 75% alcohol for 20 sec, 10% H2O2 for 10 min, washed five times with sterile water and finally placed on MS medium supplemented with 75 mg/L kanamycin for 2–3 weeks. The survivals were then transferred into a soil mix containing peat moss/perlite (2/1, v/v) in a plastic tray and grown in a greenhouse for another 2–3 weeks. Transgenic and wild‐type tobacco plants of the same size were transferred into a soil mix containing peat moss/general soil (2/1, v/v) in plastic pots for another 3–4 weeks. Peanut and tobacco plants were grown in the greenhouse at 26 °C and 70% relative humidity under a 16 h/8 h light/dark cycle.
Pathogens and inoculation
Virulent strains Rs‐P.362200 and FJ1003 strain of R. solanacearum were from peanut and tobacco, respectively. The pathogen strains were streaked on TTC agar medium (0.5 g/L 2,3,5‐triphenyltetrazolium chloride, 5 g/L peptone, 0.1 g/L casein hydrolysate, 2 g/L D‐glucose and 15 g/L agar) (Kelman, 1954) and then incubated at 28 °C for 48 h. Virulent colonies were harvested with sterile water (with 0.02% Tween‐20), and the inoculum was prepared by adjusting the concentration of bacterial cells to an optical density of 0.5 at 600 nm wavelength (NanoDrop 2000c; Thermo Fisher Scientific, San Jose, CA, USA), corresponding to approximately 108 cfu/mL.
Then, 4‐week‐old peanut seedlings of Yueyou 92 and Xinhuixiaoli were inoculated at the third and fourth leaves from the upperpart by leaflet cutting (perpendicular to the midrib of leaflet, 2/3 deep cut to the midrib), and four leaflets were inoculated per plant. Control plants were inoculated with distilled water containing 0.02% Tween‐20. Two uncut leaflets of the treated leaves were harvested at the indicated time points for future analysis.
Tobacco was inoculated by infiltrating 10 μL of R. solanacearum suspension with 108 cfu/mL concentration into the third leaves from the upperpart using a syringe with a needle, and then, the fourth leaves were harvested at the indicated time points for future analysis. The typical symptoms of bacterial wilt were monitored daily in five disease severity ratings from 0 to 4, where 0 = no symptoms, 1 = 1/4 inoculated leaves wilted, 2 = 1/4−1/2 inoculated leaves wilted, 3 = 1/2−3/4 inoculated leaves wilted and 4 = whole plant wilted, plant death. Disease index (DI) and death ratio (DR) were calculated using the following formula: DI (%) = [∑ (ni × vi) ÷ (V × N)] × 100, DR (%) = (ni ÷ N) × 100, where ni = number of plants with the respective disease rating; vi = the disease rating; V = the highest disease rating; and N = the total number of observed plants.
Application of plant hormones and abiotic/biotic stresses
One‐month‐old peanut seedlings (Minhua 6) were sprayed with 3 mM SA, 10 μg/mL ABA, 1 mg/mL ET and 100 mM MeJA in distilled water (H2O). Control seedlings were sprayed with distilled water (H2O). The leaves of the treated seedlings were harvested at indicated time points, frozen in liquid nitrogen and then stored at −80 °C until used. Yueyou 92 and Xinhuixiaoli were used in another trial. Seven‐leaf peanut Minhua 6 plants were treated at 4 °C and 25 °C. Leaves were harvested at indicated time points after treatments. Minhua 6 plants at the seven‐leaf stage were treated by stopping and normal watering for drought stress. Leaves were harvested at different time points, frozen in liquid nitrogen and then stored at −80 °C until use. Three biological replicates were set for all stress treatments.
Full‐length cDNA cloning
The candidate gene was screened through microarray analysis with approximately 100 000 unigene probes on the basis of the available fragment sequence. The 5′‐ and 3′‐end cDNA sequences were cloned by RACE using the SMART™ RACE cloning kit (Clontech, Palo Alto, CA) in accordance with the manufacturer's instructions with minor revisions. Total RNA was extracted from the leaves of resistant peanut cultivar to R. solanacearum by the CTAB method. RACE‐F and 3′ PCR adaptor primers were joined on both ends of the cDNA. Then, 5′ RACE was generated by PCR using the primary primer set of RACE‐F primer and PRRS_1EW9‐R, followed by the reaction system: 94 °C for 5 min; 35 cycles of 30 s at 95 °C, 30 s at 60 °C and 1 min 30 s at 72 °C; and 72 °C for 10 min. Similarly, 3′ RACE was generated by the set of PRRS_1EW9_F and the 3′ PCR primer with the following PCR programme: 94 °C for 5 min; 5 cycles of 30 s at 95 °C and 2 min at 72 °C; and 30 cycles of 30 s at 95 °C, 60 °C, 30 s, 2 min at 72 °C; and 72 °C for 10 min. The RACE products were cloned and sequenced. After assembly, full‐length cDNA and DNA sequences of AhRRS5 were cloned from the reverse transcription products and genomic DNA by using the set of AhRRS5‐ FL‐F and AhRRS5‐FL‐R. All primers used in this study are listed in Table S1.
Sequence analysis and phylogenetic tree construction
AhRRS5 sequence similarity analysis was performed using BLASTN and BLASTX (http://www.ncbi.nlm.nih.gov/BLAST). Four known functional resistant proteins with close similarities were obtained from the BLASTX results. Multiple sequence alignments were performed with ClustalW2 (Data S1). A phylogenetic tree was generated using 29 resistant proteins of known function by using MEGA 5.10 (Data S3).
Subcellular localization
The full‐length AhRRS5 ORF without the termination codon was amplified by high‐fidelity PCR polymerase with gene‐specific primers AhRRS5‐BamH1‐F and AhRRS5‐Asc1‐R harbouring BamHI and AscI sites, respectively. The PCR products were inserted into the vector pBI‐GFP between BamHI and AscI and formed a construct with the p35S::AhRRS5‐GFP fusion gene. With pBI‐GFP containing 35S::GFP as a control, p35S::AhRRS5‐GFP and p35S::GFP were transformed into Agrobacterium tumefaciens strain GV3101. which was cultured in induction medium (10 mM ethanesulfonic acid, pH 5.7, 10 mM MgCl2 and 200 mM acetosyringone), harvested and diluted to OD600 = 0.8, and then injected into Nicotiana benthamiana leaves using a syringe without a needle. Forty‐eight hours after agroinfiltration, GFP fluorescence was imaged in a fluorescence microscope, with an excitation wavelength of 488 nm and a 505–530 nm bandpass emission filter. GFP florescence was imaged using laser confocal florescence microscopy (Leica TCS SP8, Solms, Germany).
Vector construction and transient expression
The complete ORF of AhRRS5 was amplified by high‐fidelity PCR polymerase with AhRRS5‐OE‐F and AhRRS5‐OE‐R primers harbouring BamHI and AscI sites, respectively. The PCR products were cloned into the modified vector pBI121‐GUSA between BamHI and AscI sites to replace the GUSA gene. The obtained vector containing AhRRS5 driven by the 2 × CaMV35S promoter was named p35S::AhRRS5. The p35S::AhRRS5 vector was transferred into Agrobacterium tumefaciens strains GV3101 and EHA105.
Agrobacterium tumefaciens strain GV3101 harbouring the p35S::AhRRS5 vector was cultured to OD600 = 1.0 in induction medium (10 mM ethanesulfonic acid, pH 5.7, 10 mM MgCl2 and 200 mM acetosyringone) and diluted to OD600 = 0.8. Th diluted culture was injected into Nicotiana benthamiana leaves using a syringe without a needle. For the DAB and trypan blue staining, the tobacco (N. benthamiana) leaf was infiltrated of AhRRS5 in a small syringe with 1.0 cm diameter, the volume was about 100 μL. For the electrolyte leakage analysis, the second leaf was infiltrated with about 1 mL agrobacterium until spread to the whole leaf. The infiltrated leaves were harvested at the indicated time points for future analysis. Three biological replicates were set for the experiment.
Tobacco transformation
N. tabacum cv. CB‐1, cv. Honghuadajinyuan were used as the host, and p35S::AhRRS5 fusion gene was transformed by the leaf‐disc method mediated by EHA105 to generate transgenic plants (Rizhsky et al., 2002). The initial transgenic T0 and T1 offspring were selected by kanamycin and confirmed by RT‐PCR to verify transgene integration. The T2 transgenic homozygous lines were obtained and used in this study.
Quantitative real‐time RT‐PCR
Total RNA was extracted from peanut, transgenic tobacco and wild‐type seedlings through CTAB extraction (Chen et al., 2015). Reverse transcription was performed with PrimeScript™ RTase (TaKaRa, Dalian, China) in accordance with the manufacturer's instructions. Real‐time PCR for the relative expression level of target genes was performed with specific primers (see Table S1 for gene‐specific primers) essentially provided for the Master cyclereprealplex (Eppendorf, Hamburg, Germany) and SYBR Premix Ex Taq II (Perfect Real Time; TaKaRa, Dalian China). Each reaction mix (20 μL) contained 10 μL of SYBR Premix ExTaq (2×), 0.2 μL of PCR forward/reverse gene‐specific primers (10 μm) and diluted cDNA (2 μL). Three experimental replicates were performed for each gene using different cDNAs synthesized from three biological replicates. The PCR programme was as follows: 95 °C for 5 min; 40 cycles of 5 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C; and 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s and 60 °C for 15 s. The specificity of amplification was confirmed by melting curve analysis after 40 cycles. The relative expression level of the target gene was calculated using the comparative CT method (2−ΔΔCT method) (Schmittgen and Livak, 2008) by normalizing the PCR threshold cycle number (Ct value) of the target gene with that of the reference gene. The Ct value was calculated as follows: ΔΔCt = (CTgene−CTactin) treat−(CTgene−CTactin)control. Ahactin was used as an internal reference to detect the relative transcript level of AhRRS5 under different treatments in peanut. Tobacco NtEF1α was used as an internal reference to detect the relative transcript levels of related defence genes after treatment with R. solanacearum between the wild‐type and transgenic tobacco plants.
Histochemical analysis and ion conductivity determination
Transient expression development was assessed 48 h after the transient overexpression of AhRRS5 in tobacco leaves by staining the infected plants with 3, 3′‐diaminobenzidine (DAB; Sigma, St. Louis, MO) and lactophenol–ethanol–trypan blue. The infected tobacco leaves were incubated in 1 mg/mL DAB solution overnight at room temperature, boiled for 5 min in a solution of 3:1:1 ethanol/lactic acid/glycerol and then placed in absolute ethanol before observation to measure H2O2 level. Cell death was detected by boiling the inoculated leaves in trypan blue staining solution (10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, 30 mL of absolute ethanol and 10 mg of trypan blue, dissolved in 10 mL of ddH2O) for 2 min. The leaves were left at room temperature overnight, transferred into chloral hydrate solution (2.5 g of chloral hydrate dissolved in 1 mL of distilled water) and then boiled for 20 min to destain. The leaves were observed under a light microscope.
Ion conductivity was measured as previously described with minor modifications (Hwang and Hwang, 2011). Six round leaf discs (11 mm in diameter) per agroinfiltrated leave were cut, washed in ddH2O and then incubated in 20 mL of ddH2O with evacuation for 10 min at room temperature. Electrolyte leakage was measured using MettlerToledo 326.
Microarray analysis
In silico analysis of AhRRS5 gene expression pattern in peanut, microarray designing, hybridization, washing, and scanning and data analysis were performed as described by Chen et al. (2015). The gene expression intensity of all hybridizations was analysed, and expression levels were estimated among different tissues and under diverse stress conditions. The expression data of genes were normalized using quantile normalization (Bolstad et al., 2003) and generated using the Robust Multichip Average algorithm (Irizarry et al., 2003a,b). Three replicates were performed for all experiments.
Tobacco microarray analysis was performed using the leaves of the hyper‐resistant tobacco variety Yanyan 97, hypersusceptible tobacco variety Honghuadajinyuan, T2 generation transgenic tobacco of AhRRS5‐OE‐3, and wild‐type tobacco after R. solanacearum inoculation. Microarray designing, hybridization, washing, and scanning and data analysis were conducted as previously described (Zhang et al., 2016). Gene expression data were analysed as follows.
Supporting information
Table S1 Main primers for PCR used in this study.
Table S2 Detailed data of disease indexes and death ratios of different OE lines and the wild type after inoculation with Ralstonia solanacearum.
Data S1 Sequences of AhRRS5 full‐length cDNA, genomic DNA and protein.
Data S2 Amino acid sequences of four homologous R genes (list the function).
Data S3 Twenty‐nine known functional R genes used for phylogenetic analysis.
Data S4 In silico study of expression characteristics of four NBS‐LRR gene members in the AhRRS5 family in peanut.
Data S5 qPCR analysis of relative transcript levels of defence marker genes in leaves of T2 AhRRS5‐OE‐3 lines compared to that in leaves of wild‐type tobacco plants.
Acknowledgement
This work was supported by the International Cooperation Program (2008DFA31450) and the National 863 Program (2013AA102602) of the Ministry of Science and Technology of P.R. China.
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis. Proc. Natl Acad. Sci. 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameline‐Torregrosa, C. , Wang, B.‐B. , O'Bleness, M.S. , Deshpande, S. , Zhu, H. , Roe, B. , Young, N.D. et al (2008) Identification and characterization of nucleotide‐binding site‐leucine‐rich repeat genes in the model plant Medicago truncatula . Plant Physiol. 146, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B. , Zambryski, P. , Staskawicz, B. and Dinesh‐Kumar, S.P. (1997) Signaling in plant‐microbe interactions. Science, 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Bari, R. and Jones, J.D.G. (2009) Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Beckers, G.J.M. and Spoel, S.H. (2006) Fine‐tuning plant defence signalling: salicylate versus jasmonate. Plant Biol. 8, 1–10. [DOI] [PubMed] [Google Scholar]
- Ben, C. , Debellé, F. , Berges, H. , Bellec, A. , Jardinaud, M.‐F. , Anson, P. , Huguet, T. et al (2013) MtQRRS1, an R‐locus required for Medicago truncatula quantitative resistance to Ralstonia solanacearum . New Phytol. 199, 758–772. [DOI] [PubMed] [Google Scholar]
- Bertioli, D.J. , Leal‐Bertioli, S.C.M. , Lion, M.B. , Santos, V.L. , Pappas, G. , Cannon, S.B. and Guimarães, P.M. (2003) A large scale analysis of resistance gene homologues in Arachis. Mol. Genet. Genomics, 270, 34–45. [DOI] [PubMed] [Google Scholar]
- Bhatnagar‐Mathur, P. , Sunkara, S. , Bhatnagar‐Panwar, M. , Waliyar, F. and Sharma, K.K. (2015) Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci. 234, 119–132. [DOI] [PubMed] [Google Scholar]
- Bolstad, B.M. , Irizarry, R.A. , Åstrand, M. and Speed, T.P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics, 19, 185–193. [DOI] [PubMed] [Google Scholar]
- Bonardi, V. , Tang, S. , Stallmann, A. , Roberts, M. , Cherkis, K. and Dangl, J.L. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl Acad. Sci. 108, 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C. , Nam, J. and Dangl, J.L. (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl Acad. Sci. 95, 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert, W.F. , Delauré, S.L. , De Bolle, M.F.C. and Cammue, B.P.A. (2006) The role of ethylene in host‐pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416. [DOI] [PubMed] [Google Scholar]
- Brogue, K. , Chet, I. , Holliday, M. , Cressman, R. , Biddle, P. , Knowlton, S. , Mauvais, C.J. et al (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani . Science, 254, 1194–1197. [DOI] [PubMed] [Google Scholar]
- Browse, J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. [DOI] [PubMed] [Google Scholar]
- Carmeille, A. , Caranta, C. , Dintinger, J. , Prior, P. , Luisetti, J. and Besse, P. (2006) Identification of QTLs for Ralstonia solanacearum race 3‐phylotype II resistance in tomato. Theor. Appl. Genet. 113, 110–121. [DOI] [PubMed] [Google Scholar]
- Cesari, S. , Thilliez, G. , Ribot, C. , Chalvon, V. , Michel, C. , Jauneau, A. , Rivas, S. et al (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR‐Pia and AVR1‐CO39 by direct binding. Plant Cell, 25, 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N. , Goodwin, P.H. and Hsiang, T. (2003) The role of ethylene during the infection of Nicotiana tabacum by Colletotrichum destructivum . J. Exp. Bot. 54, 2449–2456. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Shi, Y. , Liu, W. , Chai, R. , Fu, Y. , Zhuang, J. and Wu, J. (2011) A Pid3 allele from rice cultivar Gumei2 confers resistance to Magnaporthe oryzae . J. Genet. Genomics, 38, 209–216. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Zhang, C. , Deng, Y. , Zhou, S. , Zheng, Y. , Ma, S. , Tang, R. et al (2016) Identification of low Ca2 + stress‐induced embryo apoptosis response genes in Arachis hypogaea by SSH‐associated library lift (SSHaLL). Plant Biotechnol. J. 14, 682–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh, D. , Aarons, S. , McGill, G.E. and Young, N.D. (1994) Genetic dissection of oligogenic resistance to bacterial wilt in tomato. Mol. Plant Microbe Interact. 7, 464–471. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. , Dietrich, R.A. and Richberg, M.H. (1996) Death don't have no mercy: cell death programs in plant‐microbe interactions. Plant Cell, 8, 1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, B. , Dahlbeck, D. and Staskawicz, B.J. (2006) NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis . Plant Cell, 18, 2782–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer, D. , Van Buyten, E. , Satoh, K. , Balidion, J. , Mauleon, R. , Choi, I.‐R. , Vera‐Cruz, C. et al (2012) Brassinosteroids antagonize gibberellin‐and salicylate‐mediated root immunity in rice. Plant Physiol. 158, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Theulières, F. , Hirsch, J. , Feng, D.X. , Bittner‐Eddy, P. , Beynon, J. et al (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1‐R gene, a member of a novel family of resistance genes. Proc. Natl Acad. Sci. 99, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, I. et al (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi, U.K. , Rahman, T. and Krishna, P. (2010) Brassinosteroid‐mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. (1998) SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. and Somssich, I.E. (2007) Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gabriëls, S.H.E.J. , Vossen, J.H. , Ekengren, S.K. , van Ooijen, G. , Abd‐El‐Haliem, A.M. , Berg, G. , Rainey, D.Y. et al (2007) An NB‐LRR protein required for HR signalling mediated by both extra‐and intracellular resistance proteins. Plant J. 50, 14–28. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Xu, Z. , Jiao, F. , Yu, H. , Xiao, B. , Li, Y. and Lu, X. (2010) Cloning, structural features, and expression analysis of resistance gene analogs in tobacco. Mol. Biol. Rep. 37, 345–354. [DOI] [PubMed] [Google Scholar]
- Gao, Z. , Chung, E.‐H. , Eitas, T.K. and Dangl, J.L. (2011) Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl Acad. Sci. 108, 7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Godiard, L. , Sauviac, L. , Torii, K.U. , Grenon, O. , Mangin, B. , Grimsley, N.H. and Marco, Y. (2003) ERECTA, an LRR receptor‐like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36, 353–365. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1997) Programmed cell death in plant‐pathogen interactions. Annu. Rev. Plant Biol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Gururani, M.A. , Venkatesh, J. , Upadhyaya, C.P. , Nookaraju, A. , Pandey, S.K. and Park, S.W. (2012) Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 78, 51–65. [Google Scholar]
- Henry, E. , Yadeta, K.A. and Coaker, G. (2013) Recognition of bacterial plant pathogens: local, systemic and transgenerational immunity. New Phytol. 199, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández‐Blanco, C. , Feng, D.X. , Hu, J. , Sánchez‐Vallet, A. , Deslandes, L. , Llorente, F. , Berrocal‐Lobo, M. et al (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell Online, 19, 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S.H. , Webb, C.A. , Smith, S.M. and Sun, Q. (2001) Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312. [DOI] [PubMed] [Google Scholar]
- Hwang, I.S. and Hwang, B.K. (2011) The pepper mannose‐binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 155, 447–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A. , Bolstad, B.M. , Collin, F. , Cope, L.M. , Hobbs, B. and Speed, T.P. (2003a) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A. , Hobbs, B. , Collin, F. , Beazer‐Barclay, Y.D. , Antonellis, K.J. , Scherf, U. , Speed, T.P. et al (2003b) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Jiang, C.‐J. , Shimono, M. , Sugano, S. , Kojima, M. , Yazawa, K. , Yoshida, R. , Inoue, H. et al (2010) Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice‐Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 23, 791–798. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Keen, N.T. (1990) Gene‐for‐gene complementarity in plant‐pathogen interactions. Annu. Rev. Genet. 24, 447–463. [DOI] [PubMed] [Google Scholar]
- Kelman, A. (1954) The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in a tetrazolium medium. Phytopathology, 44, 693–695. [Google Scholar]
- Keneni, G. , Bekele, E. , Imtiaz, M. and Dagne, K. (2012) Genetic vulnerability of modern crop cultivars: causes, mechanism and remedies. Int. J. Plant Res. 2, 69–79. [Google Scholar]
- Kim, M.G. (2006) The Molecular Battle between Virulence Weapons of Pseudomonas syringae and Integrated Defense Responses of Arabidopsis thaliana. Ohio: The Ohio State University. [Google Scholar]
- Koornneef, A. and Pieterse, C.M.J. (2008) Cross talk in defense signaling. Plant Physiol. 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Lahaye, T. (2002) The Arabidopsis RRS1‐R disease resistance gene‐uncovering the plant's nucleus as the new battlefield of plant defense? Trends Plant Sci. 7, 425–427. [DOI] [PubMed] [Google Scholar]
- Lamb, C.J. , Lawton, M.A. , Dron, M. and Dixon, R.A. (1989) Signals and transduction mechanisms for activation of plant defenses against microbial attack. Cell, 56, 215–224. [DOI] [PubMed] [Google Scholar]
- Lebeau, A. , Gouy, M. , Daunay, M.C. , Wicker, E. , Chiroleu, F. , Prior, P. , Frary, A. et al (2013) Genetic mapping of a major dominant gene for resistance to Ralstonia solanacearum in eggplant. Theor. Appl. Genet. 126, 143–158. [DOI] [PubMed] [Google Scholar]
- Leister, R.T. , Ausubel, F.M. and Katagiri, F. (1996) Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1 . Proc. Natl Acad. Sci. 93, 15497–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Gao, R. , Cui, R. , Lü, B. , Li, X. , Zhao, Y. , You, Z. et al (2012) Tobacco TTG2 suppresses resistance to pathogens by sequestering NPR1 from the nucleus. J. Cell Sci. 125, 4913–4922. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Liu, X. , Dai, L. and Wang, G. (2007) Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J. Genet. Genomics, 34, 765–776. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Zhang, C. , Albrecht, U. , Shimizu, R. , Wang, G. and Bowman, K.D. (2013) Overexpression of a citrus NDR1 ortholog increases disease resistance in Arabidopsis . Front. Plant Sci. 4, 10–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin, B. , Thoquet, P. , Olivier, J. and Grimsley, N.H. (1999) Temporal and multiple quantitative trait loci analyses of resistance to bacterial wilt in tomato permit the resolution of linked loci. Genetics, 151, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M. and Woffenden, B.J. (2003) Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol. 21, 178–183. [DOI] [PubMed] [Google Scholar]
- McHale, L. , Tan, X. , Koehl, P. and Michelmore, R.W. (2006) Plant NBS‐LRR proteins: adaptable guards. Genome Biol. 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C. , Kozik, A. , Griego, A. , Kuang, H. and Michelmore, R.W. (2003) Genome‐wide analysis of NBS‐LRR–encoding genes in Arabidopsis. Plant Cell Online, 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C. , Kaushik, S. and Nandety, R.S. (2005) Evolving disease resistance genes. Curr. Opin. Plant Biol. 8, 129–134. [DOI] [PubMed] [Google Scholar]
- Mindrinos, M. , Katagiri, F. , Yu, G.‐L. and Ausubel, F.M. (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide‐binding site and leucine‐rich repeats. Cell, 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Mur, L.A.J. , Kenton, P. , Atzorn, R. , Miersch, O. and Wasternack, C. (2006) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar, K. , Kyndt, T. , Nzogela, Y.B. and Gheysen, G. (2012) Abscisic acid interacts antagonistically with classical defense pathways in rice–migratory nematode interaction. New Phytol. 196, 901–913. [DOI] [PubMed] [Google Scholar]
- Narusaka, M. , Shirasu, K. , Noutoshi, Y. , Kubo, Y. , Shiraishi, T. , Iwabuchi, M. and Narusaka, Y. (2009) RRS1 and RPS4 provide a dual resistance‐gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. [DOI] [PubMed] [Google Scholar]
- Peart, J.R. , Mestre, P. , Lu, R. , Malcuit, I. and Baulcombe, D.C. (2005) NRG1, a CC‐NB‐LRR protein, together with N, a TIR‐NB‐LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 15, 968–973. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. and Van Loon, L.C. (2004) NPR1: the spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 7, 456–464. [DOI] [PubMed] [Google Scholar]
- Qian, Y. , Wang, X. , Wang, D. , Zhang, L. , Zu, C. , Gao, Z. , Zhang, H. et al (2013) The detection of QTLs controlling bacterial wilt resistance in tobacco (N. tabacum L.). Euphytica, 192, 259–266. [Google Scholar]
- Reddy, P.P. (2016) Viral diseases and their management. Sustain. Crop Prot. Prot. Cultiv. 161–176. [Google Scholar]
- Repetti, P.P. , Day, B. , Dahlbeck, D. , Mehlert, A. and Staskawicz, B.J. (2004) Overexpression of the plasma membrane‐localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana . Plant J. 40, 225–237. [DOI] [PubMed] [Google Scholar]
- Rizhsky, L. , Liang, H. and Mittler, R. (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 130, 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A. , Neuenschwander, U.H. , Willits, M.G. , Molina, A. , Steiner, H.‐Y. and Hunt, M.D. (1996) Systemic acquired resistance. Plant Cell, 8, 1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu, D. , Tasma, I.M. , Frasch, R. and Bhattacharyya, M.K. (2009) Systemic acquired resistance in soybean is regulated by two proteins, orthologous to Arabidopsis NPR1. BMC Plant Biol. 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Selote, D. , Shine, M.B. , Robin, G.P. and Kachroo, A. (2014) Soybean NDR1‐like proteins bind pathogen effectors and regulate resistance signaling. New Phytol. 202, 485–498. [DOI] [PubMed] [Google Scholar]
- Sinapidou, E. , Williams, K. , Nott, L. , Bahkt, S. , Tör, M. , Crute, I. , Bittner‐Eddy, P. et al (2004) Two TIR: NB: LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis . Plant J. 38, 898–909. [DOI] [PubMed] [Google Scholar]
- Sohn, S.‐I. , Kim, Y.‐H. , Kim, B.‐R. , Lee, S.‐Y. , Lim, C.K. , Hur, J.H. and Lee, J.‐Y. (2007) Transgenic tobacco expressing the hrpN (EP) gene from Erwinia pyrifoliae triggers defense responses against botrytis cinerea. Mol. Cells, 24, 232. [PubMed] [Google Scholar]
- Sohn, K.H. , Segonzac, C. , Rallapalli, G. , Sarris, P.F. , Woo, J.Y. , Williams, S.J. , Newman, T.E. et al (2014) The nuclear immune receptor RPS4 is required for RRS1 SLH1‐dependent constitutive defense activation in Arabidopsis thaliana . PLoS Genet. 10, e1004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.J. , Palomino, C. , Satovic, Z. , Cubero, J.I. and Torres, A.M. (2006) Identification and characterization of NBS‐LRR class resistance gene analogs in faba bean (Vicia faba L.) and chickpea (Cicer arietinum L.). Genome, 49, 1227–1237. [DOI] [PubMed] [Google Scholar]
- Sunkara, S. , Bhatnagar‐Mathur, P. and Sharma, K.K. (2014) Transgenic interventions in peanut crop improvement: progress and prospects. Genet. Genomics Breed. Peanuts, 178, 179–216. [Google Scholar]
- Takahashi, H. , Miller, J. , Nozaki, Y. , Takeda, M. , Shah, J. , Hase, S. , Ikegami, M. et al (2002) RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 32, 655–667. [DOI] [PubMed] [Google Scholar]
- Takken, F.L.W. and Joosten, M.H.A.J. (2000) Plant resistance genes: their structure, function and evolution. Eur. J. Plant Pathol. 106, 699–713. [Google Scholar]
- Thomma, B.P.H.J. , Eggermont, K. , Penninckx, I.A.M.A. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P.A. and Broekaert, W.F. (1998) Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl Acad. Sci. 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. , Nürnberger, T. and Joosten, M.H.A.J. (2011) Of PAMPs and effectors: the blurred PTI‐ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoquet, P. , Olivier, J. , Sperisen, C. , Rogowsky, P. , Laterrot, H. and Grimsley, N. (1996) Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii7996. Mol. Plant Microbe Interact. 9, 826–836. [Google Scholar]
- Ton, J. , Flors, V. and Mauch‐Mani, B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, G. , Mayr, G. , Kasiem, M.M.A. , Albrecht, M. , Cornelissen, B.J.C. and Takken, F.L.W. (2008) Structure–function analysis of the NB‐ARC domain of plant disease resistance proteins. J. Exp. Bot. 59, 1383–1397. [DOI] [PubMed] [Google Scholar]
- Vos, I.A. , Moritz, L. , Pieterse, C.M.J. and Van Wees, S.C.M. (2015) Impact of hormonal crosstalk on plant resistance and fitness under multi‐attacker conditions. Front. Plant Sci. 6, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, H. , Yuan, W. , Ye, Q. , Wang, R. , Ruan, M. , Li, Z. , Zhou, G. et al (2012) Analysis of TIR‐ and non‐TIR‐NBS‐LRR disease resistance gene analogous in pepper, characterization, genetic variation, functional divergence and expression patterns. BMC Genom. 13, 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.‐F. , Olivier, J. , Thoquet, P. , Mangin, B. , Sauviac, L. and Grimsley, N.H. (2000) Resistance of tomato line Hawaii7996 to Ralstonia solanacearum Pss4 in Taiwan is controlled mainly by a major strain‐specific locus. Mol. Plant Microbe Interact. 13, 6–13. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Shine, M.B. , Gao, Q.‐M. , Navarre, D. , Jiang, W. , Liu, C. , Chen, Q. et al (2014) Enhanced disease susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol. 165, 1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E.R. , Payne, G.B. , Moyer, M.B. , Williams, S.C. , Dincher, S.S. , Sharkey, K.C. , Beck, J.J. et al (1991) Differential regulation of beta‐1,3‐glucanase messenger RNAs in response to pathogen infection. Plant Physiol. 96, 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, E. , Grassart, L. , Coranson‐Beaudu, R. , Mian, D. , Guilbaud, C. , Fegan, M. and Prior, P. (2007) Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol. 73, 6790–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller, L. , Zhang, Y. , Jones, J.D.G. and Parker, J.E. (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1‐dependent defense. Curr. Biol. 17, 2023–2029. [DOI] [PubMed] [Google Scholar]
- Xiao, S. and Chye, M.‐L. (2011) Overexpression of Arabidopsis ACBP3 enhances NPR1‐dependent plant resistance to Pseudomonas syringe pv tomato DC3000. Plant Physiol. 156, 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D.‐L. , Yang, Y. and He, Z. (2013) Roles of plant hormones and their interplay in rice immunity. Mol. Plant, 6, 675–685. [DOI] [PubMed] [Google Scholar]
- Yoo, S.‐D. , Cho, Y. and Sheen, J. (2009) Emerging connections in the ethylene signaling network. Trends Plant Sci. 14, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S.L. , Wang, C.T. , Yang, Q.L. , Zhang, D.X. , Zhang, X.Y. , Cao, Y.L. , Liang, X.Q. et al (2011) Peanut Genetics and Breeding in China, pp. 565 Shanghai: Shanghai Scientific and Technology Press. [Google Scholar]
- Zhang, J. and Zhou, J.M. (2010) Plant immunity triggered by microbial molecular signatures. Mol. Plant, 3, 783–793. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Pan, S. , Chen, H. , Cai, T.C. , Zhuang, C.H. , Deng, Y. , Zhuang, Y.H. et al (2016) Characterization of NtREL1, a novel root‐specific gene from tobacco, and upstream promoter activity analysis in homologous and heterologous hosts. Plant Cell Rep. 35, 757–769. [DOI] [PubMed] [Google Scholar]
- Zhao, B. , Ardales, E.Y. , Raymundo, A. , Bai, J. , Trick, H.N. , Leach, J.E. and Hulbert, S.H. (2004) The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol. Plant Microbe Interact. 17, 771–779. [DOI] [PubMed] [Google Scholar]
- Zhao, B. , Lin, X. , Poland, J. , Trick, H. , Leach, J. and Hulbert, S. (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc. Natl Acad. Sci. 102, 15383–15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Li, C. , Liu, Y. , Yan, C. , Zhang, T. , Zhuang, W. and Shan, S. (2012) Cloning and characterization of a NBS‐LRR resistance gene from peanut. J. Agric. Sci. 4, 243–252. [Google Scholar]
- Zvereva, A.S. and Pooggin, M.M. (2012) Silencing and innate immunity in plant defense against viral and non‐viral pathogens. Viruses, 4, 2578–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Main primers for PCR used in this study.
Table S2 Detailed data of disease indexes and death ratios of different OE lines and the wild type after inoculation with Ralstonia solanacearum.
Data S1 Sequences of AhRRS5 full‐length cDNA, genomic DNA and protein.
Data S2 Amino acid sequences of four homologous R genes (list the function).
Data S3 Twenty‐nine known functional R genes used for phylogenetic analysis.
Data S4 In silico study of expression characteristics of four NBS‐LRR gene members in the AhRRS5 family in peanut.
Data S5 qPCR analysis of relative transcript levels of defence marker genes in leaves of T2 AhRRS5‐OE‐3 lines compared to that in leaves of wild‐type tobacco plants.


