Abstract
B-cell malignancies (BCM) originate from the same cell of origin, but at different maturation stages and have distinct clinical phenotypes. Although genetic risk variants for individual BCMs have been identified, an agnostic, genome-wide search for shared genetic susceptibility has not been performed. We explored genome-wide association studies of chronic lymphocytic leukaemia (CLL, N = 1,842), Hodgkin lymphoma (HL, N = 1,465) and multiple myeloma (MM, N = 3,790). We identified a novel pleiotropic risk locus at 3q22.2 (NCK1, rs11715604, P = 1.60 × 10−9) with opposing effects between CLL (P = 1.97 × 10−8) and HL (P = 3.31 × 10−3). Eight established non-HLA risk loci showed pleiotropic associations. Within the HLA region, Ser37 + Phe37 in HLA-DRB1 (P = 1.84 × 10−12) was associated with increased CLL and HL risk (P = 4.68 × 10−12), and reduced MM risk (P = 1.12 × 10−2), and Gly70 in HLA-DQB1 (P = 3.15 × 10−10) showed opposing effects between CLL (P = 3.52 × 10−3) and HL (P = 3.41 × 10−9). By integrating eQTL, Hi-C and ChIP-seq data, we show that the pleiotropic risk loci are enriched for B-cell regulatory elements, as well as an over-representation of binding of key B-cell transcription factors. These data identify shared biological pathways influencing the development of CLL, HL and MM. The identification of these risk loci furthers our understanding of the aetiological basis of BCMs.
Differing in their clinical phenotype, chronic lymphocytic leukaemia (CLL), Hodgkin lymphoma (HL), and multiple myeloma (MM) are all malignancies resulting from the unrestrained clonal expansion of B-cells at different stages of maturation1,2,3,4. Evidence for inherited genetic susceptibility to CLL, HL and MM has been provided by studies of familial risk, and more recently from genome-wide association studies (GWAS) which have identified risk SNPs for each tumour type5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20. While the familial risks for CLL, HL and MM are primarily tumour-specific21,22, there is some epidemiological evidence for shared susceptibility23,24,25,26. An example of this is provided by the pattern of familial risks associated with B-cell malignancies (BCMs) in Swedish populations25.
Genetic variation at a number of loci, such as 5p15 and 8q24, have been shown to influence the risk of a number of BCM and non-haematological cancers27,28,29,30,31,32,33,34. Identifying risk loci that can have such pleiotropic effects is important for gaining insight into shared and divergent molecular basis of different tumour types.
While conventional meta-analysis provides a powerful tool for combining distinct GWAS, this approach is suboptimal in the presence of disease heterogeneity, such as when SNP associations are only manifest in a specific subset of the diseases, or have opposing effects for different diseases. To address such shortcomings in searching for pleiotropic risk SNPs for BCM, we adopted the previously validated association analysis based on subsets (ASSET) meta-analytic approach35,36. ASSET implements an agnostic analysis exploring all possible subsets of studies to identify the strongest association signal, while accounting for the multiple tests required by the subset search, as well as any shared controls between studies. In doing so, ASSET is able to identify variants that are positively and negatively associated with different diseases.
Applying this statistical procedure to six BCM GWAS (two each of CLL, HL and MM) we report the identification of a novel pleiotropic region influencing BCM risk, as well as eight non-HLA linked pleiotropic loci that have only previously been described in single GWAS. Within the HLA region, we report two novel coding variants in class II HLA proteins which have pleiotropic effects on BCM risk.
Results
Characteristics of the six GWAS are summarised in Supplementary Table 1. After applying quality control filters and imputation of GWAS data (see methods) we analysed over 10 million variants for pleiotropic associations in 7,097 BCM cases and 7,324 controls of European ancestry. Figure 1 shows a Manhattan plot of the association test results for CLL, HL and MM.
Figure 1. Manhattan plots (−log10(P)) by chromosome.
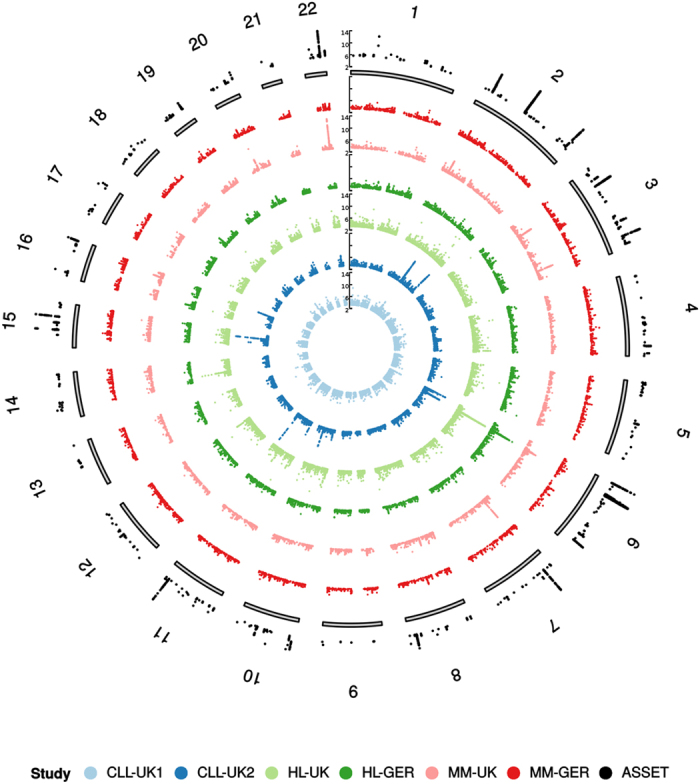
Innermost to outermost ring – chronic lymphocytic leukaemia (CLL)-UK1, CLL-UK2, Hodgkin lymphoma (HL)-UK, HL-GER, multiple myeloma (MM)-UK, MM-GER, and ASSET association test. For clarity, only data with P < 1 × 10−3 are shown.
To determine whether the global pleiotropic regions of association for pairs of BCMs occurred more often than expected by chance, we generated stratified quantile-quantile (Q-Q) plots to assess enrichment of associations for a given tumour type conditioned on the P-value for another tumour (Supplementary Fig. 1). The greater departure from the expected line associated with smaller P-values observed in the Q-Q plots provides evidence of pleiotropic effects between CLL, HL and MM37.
To identify the specific regions across the genome that demonstrate pleiotropic effects on risk of BCM we used ASSET35,36. In order for candidate SNPs to be considered, they were required to meet the following criteria: (1) variant associations at P ≤ 5.0 × 10−8 for the ASSET test; (2) at least one other variant in the same region (within r2 > 0.2) with the same pleiotropic association at P ≤ 1.0 × 10−6; (3) the individual one-sided ASSET subset tests were significant at P < 0.01; (4) the variant is not driven by a single study; (5) the variant cannot be both positively and negatively associated in different datasets of the same BCM; and (6) if a variant is positively and negatively associated with different BCM, the 2-sided P-value must be lower than both individual 1-sided P-values. Using these criteria, we identified nine non-HLA regions (607 variants).
Newly identified pleiotropic risk loci
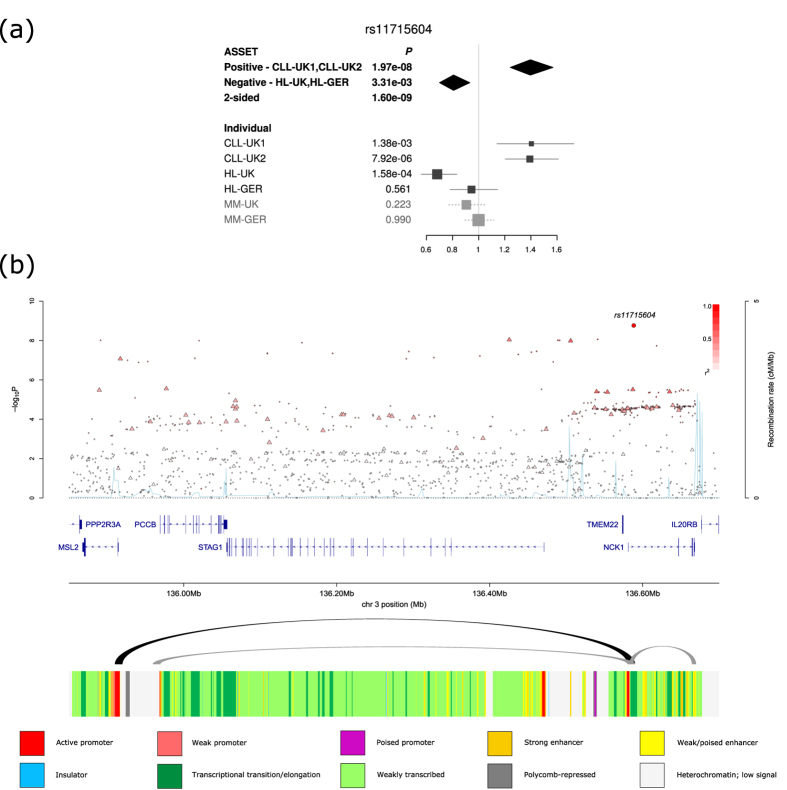
We identified a novel pleiotropic association at 3q22.2 (rs11715604, P = 1.60 × 10−9, Fig. 2) with opposing associations in CLL (P1-tailed = 1.97 × 10−8) and HL (P1-tailed = 3.31 × 10−3). rs11715604 maps to intron 1 of NCK1, which is integral to T-cell activation38,39 and regulates the PI3K/Akt pathway40. We also identified a number of promising associations that did not reach genome-wide significance, but exhibited moderate effects in the different BCM (Supplementary Table 2). These included associations at 22q13.33 (rs131821, P = 7.49 × 10−8) and 18p11.31 (rs634212, P = 5.11 × 10−5). rs131821 is intronic of NCAPH2, which is important in mitotic chromosome architecture41, while rs634212 is intronic of L3MBTL4, which has been implicated as a tumour suppressor gene for breast cancer42.
Figure 2.
(a) Forest plot of the ORs for the association between rs11715604 and BCM. Studies were weighted according to the inverse of the variance of the log of the OR calculated. Horizontal lines: 95% CI. Box: OR point estimate; box area is proportional to the weight of the study. Diamond: overall summary estimate, with CI given by its width. Unbroken vertical line: null value (OR = 1.0). (b) Regional plot of association and recombination rates. −log10(P) (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The sentinel SNP is shown as a large circle. The colour intensity of each symbol reflects the extent of LD with the sentinel SNP: white (r2 = 0) through to dark red (r2 = 1.0). Genetic recombination rates, estimated from the 1000 Genomes Project, are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to the region of association. The arcs represent Hi-C promoter contacts in GM12878 cells. The colour intensity of each contact reflects the interaction score. The bottom track represents the chromatin-state segmentation track (ChromHMM) for lymphoblastoid cells using data from the HapMap ENCODE Project.
Previously known risk loci with newly identified pleiotropic effects
We identified genome-wide significant pleiotropic associations (i.e. P ≤ 5.0 × 10−8) at eight non-HLA linked loci previously identified as risk factors for CLL, HL or MM (Table 1). The CLL risk loci at 6p21.32 (BAK1) and 6p25.3 (IRF4) were positively associated with HL risk11,17. In contrast the 2q13 (BCL2L11) and 11q24.1 (GRAMD1B) risk loci for CLL negatively influenced MM risk5,11. The MM risk locus at 3p22.1 (ULK4) positively influenced HL7, whereas the 2p23.3 (DTNB) risk locus for MM negatively associated with CLL7. The HL risk locus at 3p24.1 (EOMES) was positively associated with CLL14. The 3q26.2 (TERC) a risk factor for MM and CLL, showed a positive association with HL risk10,16. In addition to variation at these eight regions we observed promising pleiotropic associations at 2q37.1 (SP110, rs150468793; rs149207840)9, 3q27 (LPP, rs4459895)6, 5q15 (ELL2, rs2546191)18, 8q24.21 (PVT1, rs2720680)12, 15q15.1 (BMF, rs35603048)5, and 16q24.2 (IRF8, rs4240807)9 (Table 1).
Table 1. Novel pleiotropic associations in genomic regions already identified through single disease genome-wide association studies.
| Locus | SNP ID | Position (bp) | Allele 1 | Allele 2 | ASSET 2-sided P-value | Disease Group 1 |
Disease Group 2 |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BCM | Odds Ratio (CI) | P-value | BCM | Odds Ratio (CI) | P-value | ||||||
| 2p23.3 | rs6546149 | 25629438 | C | G | 6.27 × 10−10 | CLL | 1.09 (1.01–1.17) | 2.14 × 10−2 | MM | 0.83 (0.78–0.88) | 1.15 × 10−9 |
| 2q13 | rs12711846 | 111856293 | A | G | 3.48 × 10−14 | CLL | 1.44 (1.31–1.58) | 6.37 × 10−14 | MM | 0.92 (0.86–0.98) | 1.53 × 10−2 |
| 3p24.1 | rs9880772 | 27777779 | G | A | 7.42 × 10−9 | CLL, HL | 1.18 (1.11–1.24) | 7.42 × 10−9 | |||
| 3p22.1 | rs6763508 | 41750989 | T | C | 7.56 × 10−12 | MM, HL | 1.22 (1.16–1.30) | 7.56 × 10−12 | |||
| 3q26.2 | rs12638862 | 169477506 | G | A | 1.88 × 10−11 | CLL, MM, HL | 1.15 (1.09–1.19) | 1.88 × 10−11 | |||
| 6p25.3 | rs9392017 | 442357 | A | G | 6.03 × 10−9 | CLL, HL | 1.22 (1.15–1.30) | 6.03 × 10−9 | |||
| 6p21.32 | rs210143 | 33546837 | T | C | 6.81 × 10−12 | CLL, HL | 1.24 (1.17–1.32) | 6.81 × 10−12 | |||
| 11q24.1 | rs4525246 | 123395246 | G | C | 3.37 × 10−14 | CLL | 1.40 (1.28–1.53) | 6.33 × 10−14 | MM | 0.93 (0.87–0.99) | 1.50 × 10−2 |
| 2q37.1 | rs150468793; rs149207840 | 231144578 | T | TCCTCCTG | 9.63 × 10−8 | CLL, MM | 1.16 (1.10–1.22) | 9.63 × 10−8 | |||
| 3q27.3 | rs4459895 | 187954414 | A | C | 1.70 × 10−7 | CLL | 1.12 (1.02–1.23) | 1.35 × 10−2 | HL | 0.76 (0.68–0.85) | 6.43 × 10−7 |
| 5q15 | rs2546191 | 95232541 | G | A | 4.15 × 10−7 | HL | 1.18 (1.07–1.29) | 5.38 × 10−4 | MM | 0.88 (0.82–0.93) | 4.14 × 10−5 |
| 8q24.21 | rs2720680 | 129115217 | A | G | 6.78 × 10−8 | HL | 1.27 (1.16–1.39) | 1.13 × 10−7 | CLL | 0.92 (0.86–0.99) | 2.9 × 10−2 |
| 15q15.1 | rs35603048 | 40391965 | C | T | 3.64 × 10−7 | HL | 1.17 (1.06–1.28) | 1.23 × 10−3 | CLL | 0.81 (0.74–0.89) | 1.58 × 10−5 |
| 16q24.2 | rs4240807 | 85985361 | A | C | 6.62 × 10−7 | CLL | 1.21 (1.11–1.32) | 1.35 × 10−5 | HL | 0.86 (0.79–0.95) | 2.70 × 10−3 |
Genome-wide significant associations are shown above, and promising associations are below. B-cell malignancies (BCM) in bold indicate the disease the SNP was previously shown to be associated. Odds ratio calculated from allele 2.
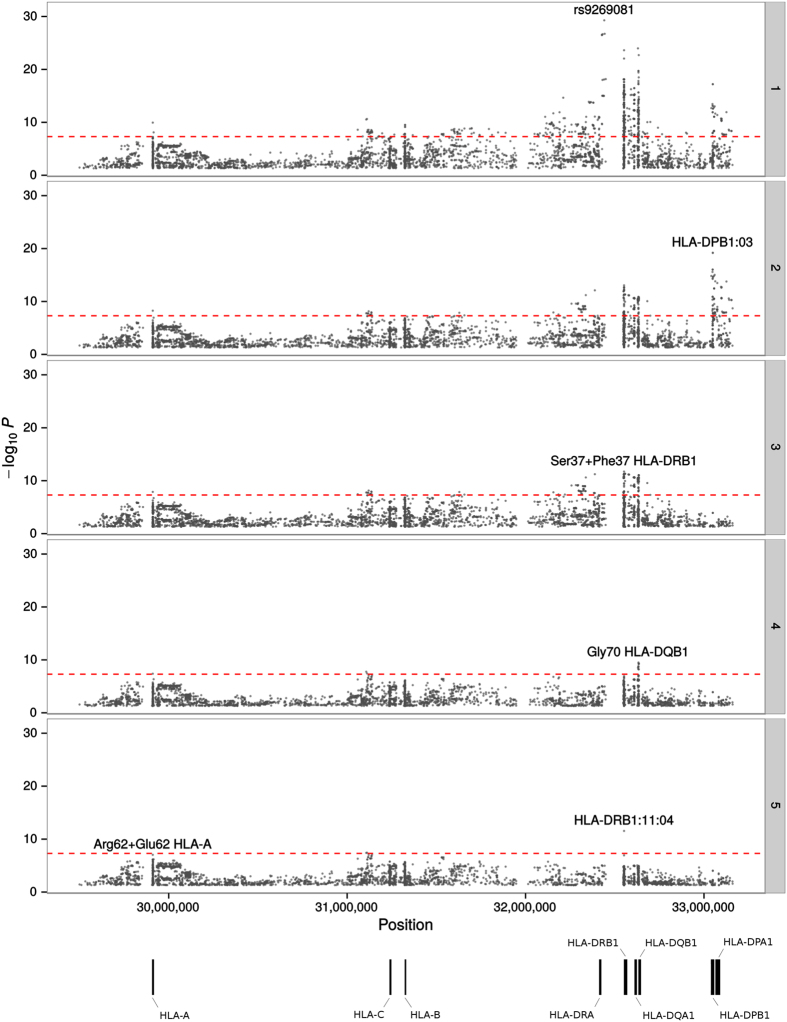
Association studies of CLL, HL and MM have demonstrated seemingly different associations between loci within the HLA region and risk10,43,44. To implement an ASSET analysis of the HLA region, we imputed classical alleles, coding variants of HLA proteins, and SNPs using the SNP2HLA software in conjunction with the Type 1 Diabetes Genetics Consortium (T1DGC) HLA reference panel45. Figure 3 shows the unconditioned ASSET associations across the 3.7 Mb HLA region. 768 variants demonstrated an association for CLL, HL and MM at P ≤ 5.0 × 10−8. To isolate independent pleiotropic associations we performed conditional stepwise logistic regression conditioning on the strongest associated variant from the 2-sided ASSET analysis. We identified Ser37 + Phe37 in HLA-DRB1 (Pconditional = 1.84 × 10−12), positively associated with CLL and HL (P1-tailed-conditional = 4.68 × 10−12) and negatively associated with MM (P1-tailed-conditional = 1.2 × 10−2). In addition, Gly70 HLA-DQB1 (Pconditional = 3.15 × 10−10) was positively associated with CLL (P1-tailed-conditional = 3.52 × 10−3) but negatively associated with HL (P1-tailed-conditional = 3.41 × 10−9). Additionally, a promising association for Arg62 + Glu62 in HLA-A (Pconditional = 9.26 × 10−8) was found, and was positively associated with CLL (P1-tailed-conditional = 8.06 × 10−5) but negatively associated with HL (P1-tailed-conditional = 5.68 × 10−5).
Figure 3. Manhattan plot representation of the step-wise conditional logistic regression of risk of BCM in the HLA region.
(1) Unconditioned test of the HLA region. (2) Results of the HLA region after conditioning on rs9269081. (3) Results of the HLA region after conditioning on rs9269081 and HLA-DPB1:03. (4) Results of the HLA region after conditioning on rs9269081, HLA-DPB1:03 and Ser37 + Phe37 HLA-DRB1. (5) Results of the HLA region after conditioning on rs9269081, HLA-DPB1:03, Ser37 + Phe37 HLA-DRB1 and Gly70 HLADQB-1. The −log10(P) of the combined logistic regression test P-values are plotted against their physical chromosomal position. The broken red line represents the genome-wide level of significance (P < 5 × 10−8).
Biological inference of pleiotropic risk loci
To explore whether the identified SNPs are eQTLs, we searched the Blood eQTL browser46, and MuTHER47 and Geuvadis/1000 Genomes48 lymphoblastoid cell line (LCL) datasets. In addition we examined expression data from MM plasma cells49. We found evidence for eQTLs (FDR adjusted P < 0.05) for nine of the pleiotropic loci in the LCL data, and four loci in the plasma cell data (Supplementary Table 3).
Since spatial proximity between specific genomic regions and chromatin looping interactions are central for regulation of gene expression50, we identified patterns of chromatin interactions at candidate pleiotropic SNPs by analysing Hi-C data on GM12878, as a source of B-cell information (Fig. 2 and Supplementary Fig. 2). Looping chromatin interactions were shown at 3q22 (rs11715604), 3p24 (rs9880772), 3q26 (rs12638862), 6p21 (rs210143) and 11q24 (rs4525246). The looping interactions at 3q22, implicates IL-20RB which regulates antigen-specific T-cell responses51. Furthermore, at 3p24, we observed looping interactions with AZI2, which contributes to the activation of NF-κB52.
Across the BCM pleiotropic risk loci, we confirmed enrichment of regulatory elements in primary haematopoietic stem cells (P = 2.1 × 10−3) and GM12878 cells (P = 7.4 × 10−3, Supplementary Table 4)53. Analysis of ChIP-seq data on 82 transcription factors (TFs) showed an enrichment of binding of key B-cell transcription factors, including CEBPB, RXRA, and POLR3G (P < 0.05/82 = 6.10 × 10−4) (Supplementary Fig. 3). CEBPB is a TF that is involved in immune and inflammatory responses54, and can induce reprogramming of B-cells into pluripotent stem cells55. RXRA can induce B-cell differentiation56, and POLR3G is a DNA-dependent RNA polymerase III57.
Pathway analysis and construction of a susceptibility network
We performed a gene-set enrichment analysis to gain insight into the biological pathways perturbed by genetic variation common to CLL, HL and MM, and found eight pathways related to the inflammatory response and antigen processing that showed enrichment (i.e. FDR adjusted P-value < 0.05; Supplementary Table 5). Following on from this analysis, we investigated the inter-connectivity of the associated genes58. By constructing a network of published and predicted protein-protein interactions, protein co-localisations and protein domain similarity, we delineated two broad clusters – one related to BCL2, and the other related to HLA (Supplementary Fig. 4).
Discussion
Motivated by the stratified Q-Q plots, which suggested the existence of pleiotropy, we utilised genotype data from six datasets in British and German populations, to conduct an agnostic cross-cancer genome-wide analysis to identify specific pleiotropic associations for CLL, HL and MM for both HLA and non-HLA regions. We identified a number of promising associations that have strong biological plausibility, including NCK1, NCAPH2 and L3MBTL4. A contemporaneous analysis also used ASSET across a number of different non-BCM cancers, and discovered a novel risk locus at 1q22 involving breast and lung cancer59.
Our analysis also provides evidence for common and opposing effects being responsible for BCM pathogenesis, but is not the first to identify opposing risk associations in different cancers60,61. Given that many of the identified risk loci harbour genes integral for immune function, it is entirely conceivable that balancing selection may act to ensure immune diversity and thus a selective advantage against temporal environmental risk factors such as infection62.
As with standard GWAS analyses ASSET may not identify the causative genetic variant at a locus. Accepting this caveat, many of the identified regions map to eQTL and regulatory elements in B-cells. Moreover, they feature an over-representation of key B-cell TF binding.
The HLA class II region has previously been implicated in multiple BCM including follicular lymphoma33,63, HL12 and CLL17,44. Here, we additionally show the involvement of this region in the development of MM. By performing a more refined imputation analysis on the HLA region, we found a variant that alters amino acid 37 of HLA-DRB1. This change affects the electrostatic properties of the P9 binding pocket64, altering T-cell receptor recognition65. The second pleiotropic association at HLA region at amino acid 70 of HLA-DQB1 is located in the P4 binding pocket, which is also a critical residue influencing antigen T-cell receptor binding66. A previous study of a number of different B-cell lymphomas using over 7,000 cases also found an association in the HLA region67, further highlighting the importance of this region to the development of BCM. In addition to the HLA association, we identified other associations that were independently ascertained in the BCM specific GWAS, including 3p24.1 (EOMES) for HL14 and CLL6, and 3q26.2 (TERC) for MM10 and CLL16, thus adding confidence that ASSET method is able to identify common genetic components.
Although predicated on protein-protein interactions, our pathway analysis provides two core cellular functions influencing BCM susceptibility. Firstly, antigen presentation/T-cell regulation, centred around HLA, and secondly cellular growth and apoptosis, centred around BCL2, which are interconnected through the key B-cell regulators, MYB and GATA368,69. BCL2 is commonly overexpressed in BCM and is relevant to tumour escape apoptosis70,71,72. It is noteworthy that Venetoclax, a BCL2 inhibitor used in treatment of CLL73, may also be efficacious in treating other forms of BCM74. This exemplifies that targeting pathways identified through GWAS may inform drug discovery initiatives75.
In conclusion, using data from six GWAS we have identified associations with multiple BCM. There are likely additional loci that have an effect, but their detection will require additional efforts with larger datasets. Such future analyses should also address the disparity in sample sizes of each of the BCM series that characterises our study.
Methods
Subjects and GWAS datasets
We used data generated from GWAS of CLL, HL, and MM performed in European populations which have been the subject of previous publications10,11,12,14,16. Briefly, the MM-UK GWAS comprised 2,282 cases (1,060 male; mean age at diagnosis: 64 years) recruited through the UK Medical Research Council (MRC) Myeloma-IX and Myeloma-XI trials. The MM-GER GWAS comprised 1,508 cases (867 male; mean age at diagnosis: 59 years) recruited by the German Multiple Myeloma Study Group (GMMMG) coordinated by the University Clinic, Heidelberg. The HL-UK GWAS comprised 622 cases ascertained through: (i) the Royal Marsden Hospital National Health Service Trust Family History study during 2004–2008 (n = 104, 63 male; mean age at diagnosis: 38 years); and (ii) an ongoing national study of HL in females (n = 518, mean age at diagnosis: 23 years) conducted by the Institute of Cancer Research (ICR). The HL-GER GWAS comprised 1,001 HL cases (597 male; mean age at diagnosis: 35 years) ascertained by the German Hodgkin Study Group during 1998–2007. The CLL-UK1 GWAS comprised 517 cases: (i) 155 cases (95 male; mean age at diagnosis: 59 years) from ICLLLC; and (ii) 362 cases (269 male; mean age at diagnosis: 63 years) from the Leukaemia Research CLL-4 trial. CLL-UK2 comprised 1,403 cases collected from two ongoing initiatives: (i) 1,111 cases collected through a UK national study of CLL genetics coordinated by the ICR (712 male; mean age at diagnosis: 63 years); and (ii) 292 cases collected through the Newcastle CLL Consortium (181 male; mean age at diagnosis: 66 years) from patients attending six haematology units in the UK.
Collection of blood samples and clinical information from subjects was undertaken with informed written consent and relevant ethical review board approval at respective institutions, in accordance with the tenets of the Declaration of Helsinki. Specifically, approval for the CLL data was approved by the UK Multi-Research Ethics Committee (MREC 99/1/082). For the MM data, the Myeloma-IX trial was approved by the Medical Research Council Leukaemia Data Monitoring and Ethics committee (MREC 02/8/95, ISRCTN68454111), the Myeloma-XI trial by the Oxfordshire Research Ethics Committee (MREC 17/09/09, ISRCTN49407852), and the GMMMG study was approved by the University of Heidelberg Ethical Commission (229/2003, S-337/2009, AFmu-119/2010). For the HL data, approval was obtained from the Multi-Research Ethics Committee (MREC 03/1/096) for the UK data, and the Ethics Committee of the University of Cologne for the German data. All methods and experimental protocols were performed in accordance with relevant guidelines and regulations.
Genotyping of cases was performed using Illumina arrays: CLL-UK1 on 317 K array, HL-UK on 660w-Quad BeadChip, and CLL-UK2, HL-GER and all MM samples using Omni-express arrays (Illumina, San Diego, CA, US). For the UK controls, we used publicly accessible data generated by the Wellcome Trust Case Control Consortium (WTCCC), the 1958 Birth Cohort (also known as the National Child Development Study) and UK Blood Service76. Genotyping of both sets of controls was conducted using Illumina Human 1.2M-Duo Custom_v1 Array BeadChips. For the German studies we utilised controls from the Heinz Nixdorf Recall study genotyped using Illumina OmniExpress array77.
Full details of the genotyping of cases and quality control can be found in previously published work10,14,16. Briefly, general genotyping quality control assessment was as previously described78 and all SNPs presented in this study passed the required thresholds. Duplicate samples were used to check genotyping quality. SNPs and samples with <95% SNPs genotyped were eliminated from the analyses. Genotype frequencies at each SNP were tested for deviation from the Hardy–Weinberg equilibrium and rejected at P < 10−5. The number of samples and variants that passed quality control is provided in Supplementary Table 1.
We have previously confirmed an absence of systematic genetic differences between cases and controls10,14,16. Prediction of the untyped SNPs was carried out using IMPUTEv2 based on a merged reference panel from UK10K (April 2014 release) and from the 1000 Genomes Project (phase 1 v3)79,80. Association meta-analyses only included markers with info scores > 0.4, imputed call rates/SNP > 0.9 and MAFs > 0.005.
Statistical analysis
The association between variants with cancer risk in each of the six GWAS was evaluated by logistic regression under a log additive model using SNPTEST v279. In the MM-GER study, genomic inflation due to population stratification was detected (λ > 1.1), so the per-allele odds ratios (ORs) were adjusted using principal components obtained from smartPCA81.
To investigate pleiotropy globally, we generated stratified Q-Q plots of association signals in one cancer stratified by the P-values in a second cancer37, for every combination of BCM. Leftward inflation in the null line is indicative of a higher degree of pleiotropy between the two tumours than expected by chance.
Subset meta-analysis was conducted using the R statistical package ASSET (association analysis based on subsets) which explores all possible subsets of “non-null” studies to identify the strongest association signal and then evaluates the significance of the signal while accounting for multiple tests required by the subset search35,36. One-tailed tests are subsequently combined to produce a 2-sided test statistic. Although ASSET has the advantage of accounting for subsets of studies with no effects and/or effects in opposing directions, where a large majority of effects are in one direction it will have lower power compared to the conventional fixed-effect analysis. The number of overlapping subjects in the GWAS (i.e. controls from WTCCC76 and Heinz-Nixdorf77 controls) were used as a covariate when estimating standard errors35. Imputed SNPs that showed significant associations were genotyped using standardised Sanger sequencing methods to confirm the imputation fidelity.
HLA imputation and analysis
To determine whether specific coding variants within HLA genes contributed to the diverse association signals, we imputed the classical HLA alleles (A, B, C, DQA1, DQB1, DRB1) and coding variants across the HLA region using SNP2HLA45. The imputation was based on a reference panel from the Type 1 Diabetes Genetics Consortium (T1DGC) consisting of genotype data from 5,225 individuals of European descent with genotyping data of 8,961 common SNPs and indel polymorphisms across the HLA region, and four digit genotyping data of the HLA class I and II molecules. This reference panel has been used previously and showed high imputation quality for the HLA region in other studies45,82,83.
To identify independent effects, dependency analyses by step-wise logistic regression were carried out by conditioning on the strongest association signal in the specific BCM. The index SNP at each region was included as a covariate, and the association statistics were recalculated for the remaining test SNPs. This process was repeated until no SNPs reached the minimum level of significance. The criteria for declaring an independent effect were defined as P < 5 × 10−8.
Functional prediction
LD between SNPs were calculated with VCFtools84 using data from the UK10K (April 2014 release) and the 1000 Genomes Project (phase 1 v3)79,80. These data were plotted using visPIG85.
To explore the epigenetic profile of genomic location associated with BCM, we used ENCODE histone modification data and HaploReg and RegulomeDB86,87 to examine whether any of the SNPs or their proxies (i.e. r2 > 0.8 in the 1000 Genomes EUR reference panel) annotate transcription factor binding or enhancer elements.
To examine enrichment in specific TF binding across risk loci we adapted the variant set enrichment method of Cowper-Sal lari et al.88 Briefly, for each risk locus, a region of strong LD (defined as r2 > 0.8 and D’ > 0.8) was determined, and these SNP were termed the associated variant set (AVS). TF ChIP-seq uniform peak data was obtained from ENCODE for the GM12878 cell line, and included data for 82 TF. For each of these marks the overlap of the SNP in the AVS and the binding sites was determined to produce a mapping tally. SNPs with the same LD structure as the risk associated SNP were randomly selected to calculate a null mapping tally. A null distribution was produced by repeating this process 10,000 times, and approximate P-values were calculated as the proportion of permutations where the null mapping tally was greater or equal to the AVS mapping tally. An enrichment score was calculated by normalising the tallies to the median of the null distribution. Thus the enrichment score is the number of standard deviations of the AVS mapping tally from the mean of the null distribution tallies.
eQTL analysis
The presence of potential eQTL was investigated through the use of several public data sets, namely the Blood eQTL browser46 in whole blood, and MuTHER47 and Geuvadis/1000 Genomes48 in lymphoblastoid cell lines (LCL).
For myeloma plasma cell eQTL analysis, we included a German (n = 658) and a UK (n = 183) case series which had been the subject of a previous eQTL analysis49 and 608 cases of a recently published US GWAS 13. Gene expression profiling of CD138-purified plasma cells using Affymetrix U133 2.0 plus arrays was performed as described89,90,91. Pre-processing of expression data was done as previously published49. Briefly, we used the Affymetrix U133 2.0 plus array custom (CDF) (v17) mapping to Entrez genes92 as chip definition file and excluded microarray probes binding to polymorphic sites. Expression data were normalized using GC-RMA. We only included genes with log2 expression > 3.5 in at least 95% of samples of each set. After quality control and excluding autosomal genes, expression data for 8,505 genes was available. The filtered set was analysed using probabilistic estimation of expression residuals (PEER)93 to infer known and hidden intervening variables, such as cytogenetic subgroups.
For the Geuvadis and MM plasma cell data, the relationship between SNPs and expression of genes located within 1 Mb was analysed using the Matrix eQTL94 package under a linear model. In all the datasets, SNPs in LD (r2 > 0.8) with the potential pleiotropic associations were explored, and were included where FDR adjusted P < 0.05.
Network analysis
Pathway enrichment analysis was performed using the Improved Gene Set Enrichment Analysis for Genome-wide Association Study (i-GSEA4GWAS v2)95. This tool also performed a functional annotation analysis on these pathways by identifying the top SNPs that map to the pathway genes, and determining if any of these SNPs fall within ENCODE peak data, namely DNase-seq peaks of open chromatin, FAIRE peaks of open chromatin, TFBS SPP-based peaks, TFBS PeakSeq-based peaks and Histone peaks. In addition, eQTLs were determined using several eQTL databases, namely eQTL Browser, GTEx and seeQTL. Common networks were identified using GeneMANIA58. This database collated data on protein and genetic interactions from a number of sources, including BioGRID, InterPro, Reactome, and Ensembl.
Promoter capture Hi-C data
To map risk SNP to interaction involving promoter contacts and identify genes involved in HL susceptibility, we analysed previously published promoter capture Hi-C data on the GM12878 cell line as a model B-cell96. The promoter capture Hi-C interactions were used to functionally annotate GWAS SNPs to seek for evidences of looping between the SNPs and the promoters of nearby genes. Reads from technical replicates were combined before processing and valid pairs were identified using HICUP97. Two biological replicates were analysed to assure reproducibility and significant interactions were determined using CHiCAGO98.
Additional Information
How to cite this article: Law, P. J. et al. Genome-wide association analysis of chronic lymphocytic leukaemia, Hodgkin lymphoma and multiple myeloma identifies pleiotropic risk loci. Sci. Rep. 7, 41071; doi: 10.1038/srep41071 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Bloodwise provided principal funding for this study in the UK (10021/05001/06002/13044). Additional funding was provided by Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund), Myeloma UK, Wellcome Trust (076113 and 085475), the Arbib Fund, and the NHS through the Biological Research Centre of the National Institute for Health Research at the Royal Marsden Hospital NHS Trust. A.S. is supported by a clinical fellowship from Cancer Research UK. Specifically, regarding the Hodgkin lymphoma UK dataset, sample and data acquisition was supported by Breast Cancer Now and the European Union. This study made use of genotyping data from the 1958 Birth Cohort. Genotyping data on controls were generated by the Wellcome Trust Sanger Institute. A full list of the investigators who contributed to the generation of the data is available at http://www.wtccc.org.uk. In Germany (Heidelberg), funding was provided by Dietmar-Hopp-Stiftung Walldorf, the University Hospital Heidelberg, Deutsche Krebshilfe, Multiple Myeloma Research Foundation and the Systems Medicine funding from the German Ministry of Education and Science. The GWAS made use of genotyping data from the population-based HNR study. The HNR study is supported by the Heinz Nixdorf Foundation (Germany). Additionally, the study is funded by the German Ministry of Education and Science and the German Research Council (DFG; projects SI 236/8-1, SI236/9-1, ER 155/6-1 and DFG CRU 216). The genotyping of the Illumina HumanOmni-1 Quad BeadChips of the HNR subjects was financed by DZNE, Bonn. We are grateful to all the patients and investigators at the individual centres for their participation. We thank the staff of the Clinical Trials Research Unit University of Leeds, the National Cancer Research Institute Haematology Clinical Studies Group and the German Multiple Myeloma Group (GMMG) secretary and investigators.
Footnotes
Author Contributions P.J.L., A.S., J.S.M., M.H. and R.S.H. designed the study. R.S.H. and G.M. obtained financial support in the UK, K.H. and H.G. obtained support in Germany. P.J.L., A.S., J.S.M. and R.S.H. drafted the manuscript. P.J.L., A.S., J.S.M. and M.H. performed the principal statistical and bioinformatic analysis. G.O. and O.L. performed further bioinformatics analyses. P.B. coordinated UK laboratory analysis and H.E.S performed sample processing. A.S. performed Sanger sequencing. G.J.M. performed ascertainment and collection of case study samples for MM-UK GWAS. D.C.J. managed and prepared Myeloma IX and XI case study DNA samples. M.K. oversaw MM-UK GWAS data processing. H.G., N.W., B.C., M.I.d.S.F., H.T., K.H. and A.F. coordinated and managed the German DNA samples and data. P.H. and M.M.N. performed and coordinated the GWAS of German cases and controls. K.-H.J. and L.E. ascertained and managed the HNR sample. N.W. performed eQTL analysis on plasma cells. A.J.S. provided samples for HL-UK GWAS, R.C. provided data on samples for HL-UK GWAS. P.H. and M.M.N. were responsible for GER-HL GWAS analysis; E.P.v.S. and A.E. were responsible for German HL patient samples. D.C. performed recruitment of CLL samples. For Newcastle, J.M.A. and D.J.A. conceived of the Newcastle CLL Consortium; J.M.A. obtained financial support, supervised laboratory management, genotyping of cases and data handling; N.J.S. and T.R. performed sample management of cases and data handling; and G.H.J., G.S., R.J.H., A.R.P., D.J.A., J.C., J.R.B., G.P., C.P. and C.F. developed protocols for recruitment of individuals with CLL and sample acquisition and performed sample collection of cases. All authors contributed to the final paper.
References
- Rickert R. C. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol 13, 578–591 (2013). [DOI] [PubMed] [Google Scholar]
- Shaffer A. L., Rosenwald A. & Staudt L. M. Lymphoid Malignancies: the dark side of B-cell differentiation. Nat Rev Immunol 2, 920–933 (2002). [DOI] [PubMed] [Google Scholar]
- Craig F. E. & Foon K. A. Flow cytometric immunophenotyping for hematologic neoplasms. Blood 111, 3941–3967 (2008). [DOI] [PubMed] [Google Scholar]
- Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer 5, 251–262 (2005). [DOI] [PubMed] [Google Scholar]
- Berndt S. I. et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet 45, 868–76 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt S. I. et al. Meta-analysis of genome-wide association studies discovers multiple loci for chronic lymphocytic leukemia. Nat Commun 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick P. et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nat Genet 44, 58–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen W. et al. A meta-analysis of Hodgkin lymphoma reveals 19p13.3 TCF3 as a novel susceptibility locus. Nat Commun 5, 3856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther-Swanepoel D. et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet 42, 132–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb D. et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat Genet 45, 1221–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bernardo M. C. et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet 40, 1204–10 (2008). [DOI] [PubMed] [Google Scholar]
- Enciso-Mora V. et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat Genet 42, 1126–30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson S. W. et al. Genome-wide scan identifies variant in 2q12.3 associated with risk for multiple myeloma. Blood 124, 2001–2003 (2014). [DOI] [PubMed] [Google Scholar]
- Frampton M. et al. Variation at 3p24.1 and 6q23.3 influences the risk of Hodgkin’s lymphoma. Nat Commun 4, 2549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J. S. et al. Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Nat Commun 7, 12050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speedy H. E. et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat Genet 46, 56–60 (2014). [DOI] [PubMed] [Google Scholar]
- Slager S. L. et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood 120, 843–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B. et al. Variants in ELL2 influencing immunoglobulin levels associate with multiple myeloma. Nat Commun 6, 7213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urayama K. Y. et al. Genome-wide association study of classical Hodgkin lymphoma and Epstein-Barr virus status-defined subgroups. J Natl Cancer Inst 104, 240–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold N. et al. The CCND1 c.870G > A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nat Genet 45, 522–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin L. R., Bjorkholm M., Kristinsson S. Y., Turesson I. & Landgren O. Highly increased familial risks for specific lymphoma subtypes. Br J Haematol 146, 91–4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristinsson S. Y. et al. Patterns of hematologic malignancies and solid tumors among 37,838 first-degree relatives of 13,896 patients with multiple myeloma in Sweden. Int J Cancer 125, 2147–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin L. R., Bjorkholm M., Kristinsson S. Y., Turesson I. & Landgren O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin’s lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica 94, 647–53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin L. R. et al. Familial aggregation of Hodgkin lymphoma and related tumors. Cancer 100, 1902–8 (2004). [DOI] [PubMed] [Google Scholar]
- Goldin L. R., Pfeiffer R. M., Li X. & Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood 104, 1850–4 (2004). [DOI] [PubMed] [Google Scholar]
- Landgren O. et al. Risk of plasma cell and lymphoproliferative disorders among 14621 first-degree relatives of 4458 patients with monoclonal gammopathy of undetermined significance in Sweden. Blood 114, 791–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerhan J. R. et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet 46, 1233–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode E. L. et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet 42, 874–879 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemeney L. A. et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet 40, 1307–12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 40, 310–5 (2008). [DOI] [PubMed] [Google Scholar]
- Tomlinson I. et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 39, 984–8 (2007). [DOI] [PubMed] [Google Scholar]
- Turnbull C. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 42, 504–507 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibola C. F. et al. Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am J Hum Genet 95, 462–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpin B. M. et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet 46, 994–1000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S. et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet 90, 821–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F. et al. Common genetic variants in the 9p21 region and their associations with multiple tumours. Br J Cancer 108, 1378–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork A. J. et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet 9, e1003449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoenkam J. et al. Non-overlapping functions of Nck1 and Nck2 adaptor proteins in T cell activation. Cell Commun Signal 12, 21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiemwattana I. et al. Essential role of the adaptor protein Nck1 in Jurkat T cell activation and function. Clin Exp Immunol 167, 99–107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Dusseault J. & Larose L. Nck1 depletion induces activation of the PI3K/Akt pathway by attenuating PTP1B protein expression. Cell Commun Signal 12, 71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y., Zhu H., Zhang R. & Noma K. I. A novel role for the condensin II complex in cellular senescence. Cell Cycle, 0 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addou-Klouche L. et al. Loss, mutation and deregulation of L3MBTL4 in breast cancers. Mol Cancer 9, 213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsianas L. et al. Multiple Hodgkin lymphoma-associated loci within the HLA region at chromosome 6p21.3. Blood 118, 670–4 (2011). [DOI] [PubMed] [Google Scholar]
- Di Bernardo M. C. et al. Risk of developing chronic lymphocytic leukemia is influenced by HLA-A class I variation. Leukemia 27, 255–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra H. J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45, 1238–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 44, 1084–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold N. et al. The 7p15.3 (rs4487645) association for multiple myeloma shows strong allele-specific regulation of the MYC-interacting gene CDCA7L in malignant plasma cells. Haematologica 100, e110–e113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus J. H. & Dekker J. The hierarchy of the 3D genome. Mol Cell 49, 773–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl C. et al. IL-20 receptor 2 signaling down-regulates antigen-specific T cell responses. J Immunol 182, 802–10 (2009). [DOI] [PubMed] [Google Scholar]
- Fujita F. et al. Identification of NAP1, a Regulatory Subunit of IκB Kinase-Related Kinases That Potentiates NF-κB Signaling. Molecular and Cellular Biology 23, 7780–7793 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L. D. & Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research 40, D930–D934 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. K. et al. MEKK1 plays a critical role in activating the transcription factor C/EBP-beta-dependent gene expression in response to IFN-gamma. Proc Natl Acad Sci USA 99, 7945–50 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B. et al. C/EBP[agr] poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 506, 235–239 (2014). [DOI] [PubMed] [Google Scholar]
- Garcia-Bates T. M. et al. Peroxisome proliferator-activated receptor gamma (PPARγ) ligands enhance human B cell antibody production and differentiation. Journal of immunology 183, 6903 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. & Roeder R. G. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes & Development 11, 1315–1326 (1997). [DOI] [PubMed] [Google Scholar]
- Warde-Farley D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research 38, W214–W220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehringer G. et al. Cross-cancer genome-wide analysis of lung, ovary, breast, prostate and colorectal cancer reveals novel pleiotropic associations. Cancer Res (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swafford A. D. et al. An allele of IKZF1 (Ikaros) conferring susceptibility to childhood acute lymphoblastic leukemia protects against type 1 diabetes. Diabetes 60, 1041–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurecht H. et al. Genome-wide comparative analysis of atopic dermatitis and psoriasis gives insight into opposing genetic mechanisms. Am J Hum Genet 96, 104–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T., Willis J. H. & Goldstein D. B. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet 8, 845–856 (2007). [DOI] [PubMed] [Google Scholar]
- Foo J. N. et al. Coding variants at hexa-allelic amino acid 13 of HLA-DRB1 explain independent SNP associations with follicular lymphoma risk. Am J Hum Genet 93, 167–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hov J. R. et al. Electrostatic Modifications of the Human Leukocyte Antigen-DR P9 Peptide-Binding Pocket and Susceptibility to Primary Sclerosing Cholangitis. Hepatology (Baltimore, Md.) 53, 1967–1976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T. & Obata F. Allogeneic recognition of HLA-DRB1*0406 by T cells with HLA-DRB1*0403: role of amino acid residue 37 on the beta sheet in T cell recognition. Immunobiology 195, 261–70 (1996). [DOI] [PubMed] [Google Scholar]
- Reichstetter S. et al. Mutational analysis of critical residues determining antigen presentation and activation of HLA-DQ0602 restricted T-cell clones. Hum Immunol 63, 185–93 (2002). [DOI] [PubMed] [Google Scholar]
- Vijai J. et al. Susceptibility loci associated with specific and shared subtypes of lymphoid malignancies. PLoS Genet 9, e1003220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Northrup D., Boukarabila H., Jacobsen S. E. & Allman D. Transcriptional repression of Gata3 is essential for early B cell commitment. Immunity 38, 930–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness S. A. Myb binding proteins: regulators and cohorts in transformation. Oncogene 18, 3039–46 (1999). [DOI] [PubMed] [Google Scholar]
- Del Gaizo Moore V. et al. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest 117, 112–21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M. et al. Expression of the bcl-2 gene in human multiple myeloma cell lines and normal plasma cells. Blood 79, 495–502 (1992). [PubMed] [Google Scholar]
- Rigal-Huguet F. et al. Frequent expression of the cell death-inducing gene Bax in Reed- Sternberg cells of Hodgkin’s disease. Blood 87, 2470–2475 (1996). [PubMed] [Google Scholar]
- Roberts A. W. et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. New England Journal of Medicine 374, 311–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. A., Huang D. & Roberts A. Targeting BCL2 for the Treatment of Lymphoid Malignancies. Seminars in Hematology 51, 219–227 (2014). [DOI] [PubMed] [Google Scholar]
- Nelson M. R. et al. The support of human genetic evidence for approved drug indications. Nat Genet 47, 856–860 (2015). [DOI] [PubMed] [Google Scholar]
- Power C. & Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study). Int J Epidemiol 35, 34–41 (2006). [DOI] [PubMed] [Google Scholar]
- Schmermund A. et al. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL Study. Risk Factors, Evaluation of Coronary Calcium and Lifestyle. Am Heart J 144, 212–8 (2002). [DOI] [PubMed] [Google Scholar]
- Anderson C. A. et al. Data quality control in genetic case-control association studies. Nat Protoc 5, 1564–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Howie B., Myers S., McVean G. & Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39, 906–13 (2007). [DOI] [PubMed] [Google Scholar]
- Abecasis G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A. L. & Reich D. Population structure and eigenanalysis. PLoS Genet 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B. et al. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am J Hum Genet 94, 522–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Achury J. et al. Fine mapping in the MHC region accounts for 18% additional genetic risk for celiac disease. Nat Genet 47, 577–578 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales M., Jager R., Migliorini G., Houlston R. S. & Henrion M. Y. visPIG--a web tool for producing multi-region, multi-track, multi-scale plots of genetic data. PLoS One 9, e107497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22, 1790–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L. D. & Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40, D930–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper-Sallari R. et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 44, 1191–1198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. A. et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 108, 1733–43 (2006). [DOI] [PubMed] [Google Scholar]
- Meissner T. et al. Gene expression profiling in multiple myeloma--reporting of entities, risk, and targets in clinical routine. Clin Cancer Res 17, 7240–7 (2011). [DOI] [PubMed] [Google Scholar]
- Zhan F. et al. The molecular classification of multiple myeloma. Blood 108, 2020–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- BrainArray Custom CDF.
- Stegle O., Parts L., Piipari M., Winn J. & Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 7, 500–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin A. A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Chang S., Guo L. & Wang J. I-GSEA4GWAS v2: a web server for functional analysis of SNPs in trait-associated pathways identified from genome-wide association study. Protein Cell 6, 221–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47, 598–606 (2015). [DOI] [PubMed] [Google Scholar]
- Wingett S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Research 4, 1310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biology 17, 127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.